Polymerisation force of a rigid filament bundle: diffusive interaction leads to sublinear force-number scaling
- PMID: 29410507
- PMCID: PMC5802839
- DOI: 10.1038/s41598-018-20259-7
Polymerisation force of a rigid filament bundle: diffusive interaction leads to sublinear force-number scaling
Abstract
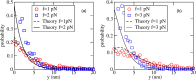
Polymerising filaments generate force against an obstacle, as in, e.g., microtubule-kinetochore interactions in the eukaryotic cell. Earlier studies of this problem have not included explicit three-dimensional monomer diffusion, and consequently, missed out on two important aspects: (i) the barrier, even when it is far from the polymers, affects free diffusion of monomers and reduces their adsorption at the tips, while (ii) parallel filaments could interact through the monomer density field ("diffusive coupling"), leading to negative interference between them. In our study, both these effects are included and their consequences investigated in detail. A mathematical treatment based on a set of continuum Fokker-Planck equations for combined filament-wall dynamics suggests that the barrier-induced monomer depletion reduces the growth velocity and also the stall force, while the total force produced by many filaments remains additive. However, Brownian dynamics simulations show that the linear force-number scaling holds only when the filaments are far apart; when they are arranged close together, forming a bundle, sublinear scaling of force with number appears, which could be attributed to diffusive interaction between the growing polymer tips.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Cooper GM. The Cell: A Molecular Approach. 2nd edition. Sunderland (MA): Sinauer Associates; 2000.
-
- Bray, D. Cell Movements: From molecules to motility (Garland Science, New York, 2001).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

