Metatranscriptome Sequencing Reveals Insights into the Gene Expression and Functional Potential of Rumen Wall Bacteria
- PMID: 29410661
- PMCID: PMC5787071
- DOI: 10.3389/fmicb.2018.00043
Metatranscriptome Sequencing Reveals Insights into the Gene Expression and Functional Potential of Rumen Wall Bacteria
Abstract
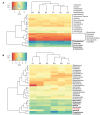
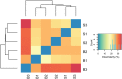
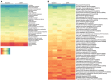
Microbiota of the rumen wall constitute an important niche of rumen microbial ecology and their composition has been elucidated in different ruminants during the last years. However, the knowledge about the function of rumen wall microbes is still limited. Rumen wall biopsies were taken from three fistulated dairy cows under a standard forage-based diet and after 4 weeks of high concentrate feeding inducing a subacute rumen acidosis (SARA). Extracted RNA was used for metatranscriptome sequencing using Illumina HiSeq sequencing technology. The gene expression of the rumen wall microbial community was analyzed by mapping 35 million sequences against the Kyoto Encyclopedia for Genes and Genomes (KEGG) database and determining differentially expressed genes. A total of 1,607 functional features were assigned with high expression of genes involved in central metabolism, galactose, starch and sucrose metabolism. The glycogen phosphorylase (EC:2.4.1.1) which degrades (1->4)-alpha-D-glucans was among the highest expressed genes being transcribed by 115 bacterial genera. Energy metabolism genes were also highly expressed, including the pyruvate orthophosphate dikinase (EC:2.7.9.1) involved in pyruvate metabolism, which was covered by 177 genera. Nitrogen metabolism genes, in particular glutamate dehydrogenase (EC:1.4.1.4), glutamine synthetase (EC:6.3.1.2) and glutamate synthase (EC:1.4.1.13, EC:1.4.1.14) were also found to be highly expressed and prove rumen wall microbiota to be actively involved in providing host-relevant metabolites for exchange across the rumen wall. In addition, we found all four urease subunits (EC:3.5.1.5) transcribed by members of the genera Flavobacterium, Corynebacterium, Helicobacter, Clostridium, and Bacillus, and the dissimilatory sulfate reductase (EC 1.8.99.5) dsrABC, which is responsible for the reduction of sulfite to sulfide. We also provide in situ evidence for cellulose and cellobiose degradation, a key step in fiber-rich feed digestion, as well as oxidative stress response and oxygen scavenging at the rumen wall. Archaea, mainly Methanocaldococcus and Methanobrevibacter, were found to be metabolically active with a high number of transcripts matching to methane and carbohydrate metabolism. These findings enhance our understanding of the metabolic function of the bovine rumen wall microbiota.
Keywords: dairy cattle; epimural microbiota; metatranscriptome; rumen epithelium; subacute rumen acidosis.
Figures

References
-
- Abrao F. O., Duarte E. R., Freitas C. E., Vieira E. A., Geraseev L. C., Da Silva-Hughes A. F., et al. (2014). Characterization of fungi from ruminal fluid of beef cattle with different ages and raised in tropical lignified pastures. Curr. Microbiol. 69 649–659. 10.1007/s00284-014-0633-5 - DOI - PubMed
-
- Abrao F. O., Duarte E. R., Pessoa M. S., Dos Santos V. L., De Freitas L. F., Barros K. D., et al. (2017). Notable fibrolytic enzyme production by Aspergillus spp. isolates from the gastrointestinal tract of beef cattle fed in lignified pastures. PLOS ONE 12:e0183628. 10.1371/journal.pone.0183628 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

