High Whole-Genome Sequence Diversity of Human Papillomavirus Type 18 Isolates
- PMID: 29414918
- PMCID: PMC5850375
- DOI: 10.3390/v10020068
High Whole-Genome Sequence Diversity of Human Papillomavirus Type 18 Isolates
Abstract
Background: The most commonly found human papillomavirus (HPV) types in cervical cancer are HPV16 and HPV18. Genome variants of these types have been associated with differential carcinogenic potential. To date, only a handful of studies have described HPV18 whole genome sequencing results. Here we describe HPV18 variant diversity and conservation of persistent infections in a longitudinal retrospective cohort study.
Methods: Cervical self-samples were obtained annually over four years and genotyped on the SPF10-DEIA-LiPA25 platform. Clearing and persistent HPV18 positive infections were selected, amplified in two overlapping fragments, and sequenced using 32 sequence primers.
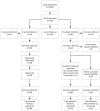
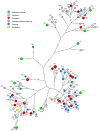
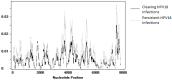
Results: Complete viral genomes were obtained from 25 participants with persistent and 26 participants with clearing HPV18 infections, resulting in 52 unique HPV18 genomes. Sublineage A3 was predominant in this population. The consensus viral genome was completely conserved over time in persistent infections, with one exception, where different HPV18 variants were identified in follow-up samples.
Conclusions: This study identified a diverse set of HPV18 variants. In persistent infections, the consensus viral genome is conserved. The identification of only one HPV18 infection with different major variants in follow-up implies that this is a potentially rare event. This dataset adds 52 HPV18 genome variants to Genbank, more than doubling the currently available HPV18 information resource, and all but one variant are unique additions.
Keywords: HPV18; genetic epidemiology; whole genome sequencing.
Conflict of interest statement
Pascal van der Weele and Audrey J. King declare no conflicts of interest. Chris J.L.M. Meijer received speaker’s fees from GlaxoSmithKline, Qiagen, SPMSD/Merck, Roche, Menarini, and Seegene. On occasion, he served on the scientific advisory boards (expert meeting) of GSK, Qiagen, SPMSD/Merck, Roche, and Genticel and as a consultant for Qiagen and Genticel. He holds a small number of Qiagen shares and is a minority shareholder of Self-Screen BV, a spin-off company of Vrije Universiteit University Medical Centre, of which he is also part time director. Until 2014, he held a small number of shares in Delphi Biosciences. Until April 2016, he was a minority stock holder of Diassay BV.
Figures

Similar articles
-
Whole-Genome Sequencing and Variant Analysis of Human Papillomavirus 16 Infections.J Virol. 2017 Sep 12;91(19):e00844-17. doi: 10.1128/JVI.00844-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28701400 Free PMC article.
-
Correlation between viral load, multiplicity of infection, and persistence of HPV16 and HPV18 infection in a Dutch cohort of young women.J Clin Virol. 2016 Oct;83:6-11. doi: 10.1016/j.jcv.2016.07.020. Epub 2016 Aug 4. J Clin Virol. 2016. PMID: 27522635
-
Molecular epidemiology of human papillomavirus 18 infections in Japanese Women.Infect Genet Evol. 2020 Sep;83:104345. doi: 10.1016/j.meegid.2020.104345. Epub 2020 Apr 28. Infect Genet Evol. 2020. PMID: 32360473
-
Genetic diversity of HPV16 and HPV18 in Brazilian patients with invasive cervical cancer.J Med Virol. 2016 Jul;88(7):1279-87. doi: 10.1002/jmv.24458. Epub 2016 Jan 12. J Med Virol. 2016. PMID: 26694554
-
Genetic signatures for lineage/sublineage classification of HPV16, 18, 52 and 58 variants.Virology. 2021 Jan 15;553:62-69. doi: 10.1016/j.virol.2020.11.003. Epub 2020 Nov 16. Virology. 2021. PMID: 33238224
Cited by
-
Genetic diversity and phylogenetic analysis of HPV 16 & 18 variants isolated from cervical specimens of women in Saudi Arabia.Saudi J Biol Sci. 2019 Feb;26(2):317-324. doi: 10.1016/j.sjbs.2018.05.005. Epub 2018 May 3. Saudi J Biol Sci. 2019. PMID: 31485171 Free PMC article.
-
Analysis of Human Papillomavirus-HPV-18 and HPV-45 Type-specific variants in a contemporary cohort of individuals with cervical disease in Ghana.Ghana Med J. 2024 Dec;58(4):311-321. doi: 10.4314/gmj.v58i4.9. Ghana Med J. 2024. PMID: 40585519 Free PMC article.
-
Abundance of HPV L1 Intra-Genotype Variants With Capsid Epitopic Modifications Found Within Low- and High-Grade Pap Smears With Potential Implications for Vaccinology.Front Genet. 2019 May 24;10:489. doi: 10.3389/fgene.2019.00489. eCollection 2019. Front Genet. 2019. PMID: 31231420 Free PMC article.
-
Fifteen new nucleotide substitutions in variants of human papillomavirus 18 in Korea : Korean HPV18 variants and clinical manifestation.Virol J. 2020 May 24;17(1):70. doi: 10.1186/s12985-020-01337-7. Virol J. 2020. PMID: 32448303 Free PMC article.
-
Genetic variability of human papillomavirus type 18 based on E6, E7 and L1 genes in central China.Virol J. 2024 Jul 5;21(1):152. doi: 10.1186/s12985-024-02424-9. Virol J. 2024. PMID: 38970084 Free PMC article.
References
-
- Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. - DOI - PubMed
-
- De Sanjose S., Quint W.G., Alemany L., Geraets D.T., Klaustermeier J.E., Lloveras B., Tous S., Felix A., Bravo L.E., Shin H.R., et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. - DOI - PubMed
-
- Xi L.F., Koutsky L.A., Hildesheim A., Galloway D.A., Wheeler C.M., Winer R.L., Ho J., Kiviat N.B. Risk for high-grade cervical intraepithelial neoplasia associated with variants of human papillomavirus types 16 and 18. Cancer Epidemiol. Biomark. Prev. 2007;16:4–10. doi: 10.1158/1055-9965.EPI-06-0670. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

