Physicochemical Properties of the Mammalian Molecular Chaperone HSP60
- PMID: 29415503
- PMCID: PMC5855711
- DOI: 10.3390/ijms19020489
Physicochemical Properties of the Mammalian Molecular Chaperone HSP60
Abstract
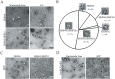
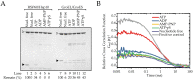
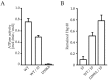
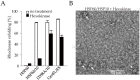
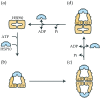
The E. coli GroEL/GroES chaperonin complex acts as a folding cage by producing a bullet-like asymmetric complex, and GroEL exists as double rings regardless of the presence of adenosine triphosphate (ATP). Its mammalian chaperonin homolog, heat shock protein, HSP60, and co-chaperonin, HSP10, play an essential role in protein folding by capturing unfolded proteins in the HSP60/HSP10 complex. However, the structural transition in ATPase-dependent reaction cycle has remained unclear. We found nucleotide-dependent association and dissociation of the HSP60/HSP10 complex using various analytical techniques under near physiological conditions. Our results showed that HSP60 exist as a significant number of double-ring complexes (football- and bullet-type complexes) and a small number of single-ring complexes in the presence of ATP and HSP10. HSP10 binds to HSP60 in the presence of ATP, which increased the HSP60 double-ring formation. After ATP is hydrolyzed to Adenosine diphosphate (ADP), HSP60 released the HSP10 and the dissociation of the double-ring to single-rings occurred. These results indicated that HSP60/HSP10 undergoes an ATP-dependent transition between the single- and double-rings in their system that is highly distinctive from the GroEL/GroES system particularly in the manner of complex formation and the roles of ATP binding and hydrolysis in the reaction cycle.
Keywords: GroEL; HSP60; chaperonin; molecular chaperone.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

