Genetic variation analysis of PCV1 strains isolated from Guangxi Province of China in 2015
- PMID: 29415728
- PMCID: PMC5803923
- DOI: 10.1186/s12917-018-1345-z
Genetic variation analysis of PCV1 strains isolated from Guangxi Province of China in 2015
Abstract
Background: Porcine circovirus type 1 (PCV1) was discovered in 1974 as a contaminant of a porcine kidney (PK-15) cell line and was generally accepted to be nonpathogenic. But recently it was shown to cause lesions in experimentally infected pig fetuses. Serological evidence and genetic studies suggested that PCV1 was widespread in domestic pigs. Thus, the molecular epidemiology and genetic variation of PCV1 are still necessary to understand.
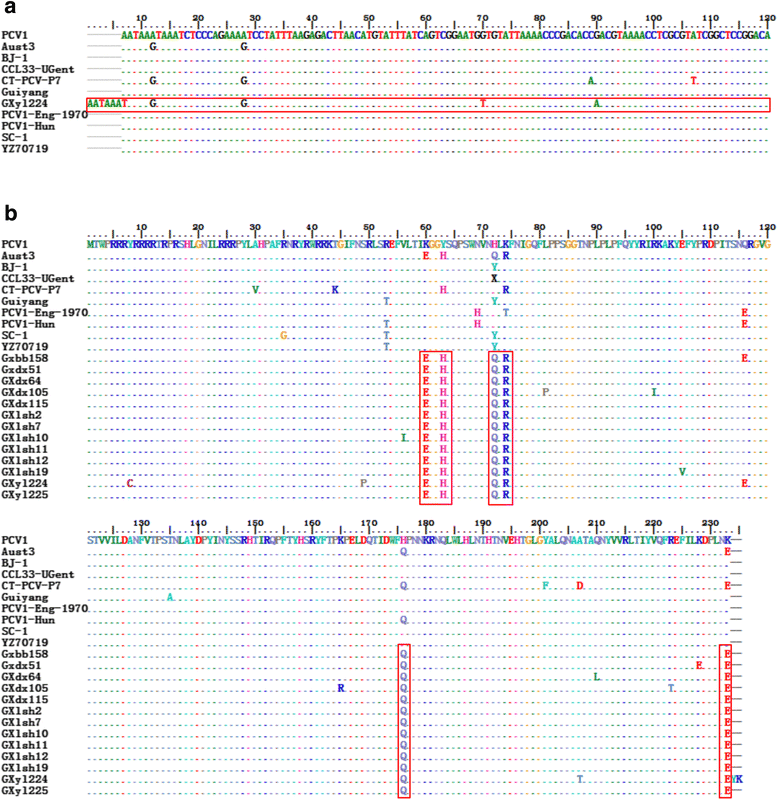
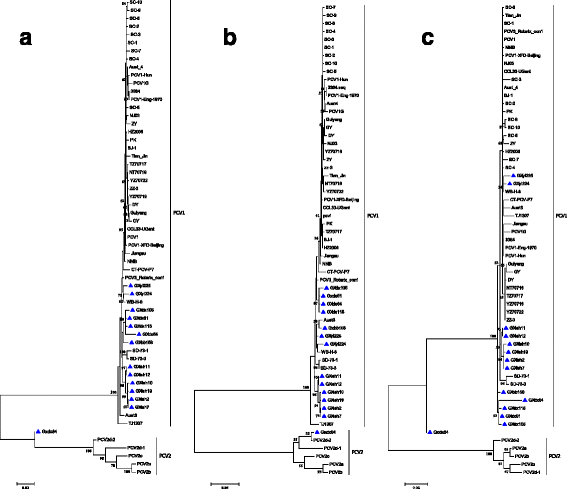
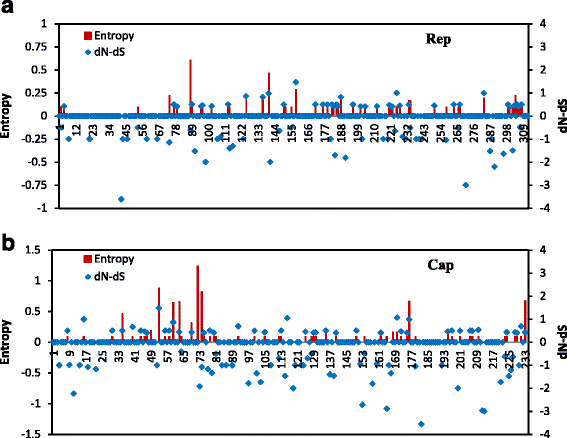
Results: Here 247 tissue samples were collected from piglets in Guangxi Province, China and performed whole-genome sequencing of the PCV1 genome. Thirteen PCV1 strains were sequenced from the samples. Similarity analysis showed that there were 97.8% to 99.6% nucleotide similarity to each other and 97.1% to 99.8% nucleotide similarity to the 40 reference strains. Besides, based on sequence analysis, we found one putative recombinant virus named GXdx84 strain contained the open-reading frame 1 (ORF1) of PCV1 and the ORF2 of PCV2d-2, which was consistent with the results of phylogenetic analysis that compared PCV1 and PCV2 strains. Variation analysis of the amino acids of the capsid protein revealed that the GXyl224 strain, which encoded 235 amino acids, had two amino acids more than other strains. This is the first study to report that a cap gene mutation resulted in lengthening of in the gene sequence.
Conclusions: These data contribute to the understanding of PCV1 evolution and molecular epidemiology that will facilitate programs for its control and prevention.
Keywords: Genetic variation; Phylogenetic study; Porcine circovirus 1; Putative recombinant virus.
Conflict of interest statement
Ethics approval and consent to participate
Experimental protocols for obtaining pigs clinical samples used in this study were obtained from Guangxi Center for Animal Disease Control and Prevention and carried out in strict accordance with the Animal Ethics Procedures and Guidelines of the People's Republic of China. All pigs were euthanized by an anesthetic overdose with the pentobarbital before collected the samples and all pig owners agree the samples’ usage before participation in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Olvera A, Cortey M, Segales J: Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology 2007, 357(2):175-185. - PubMed
-
- Tischer I, Rasch R, Tochtermann G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralblatt fur Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene Erste Abteilung Originale Reihe A: Medizinische Mikrobiologie und Parasitologie. 1974;226(2):153–167. - PubMed
-
- Allan G, Meehan B, Todd D, Kennedy S, McNeilly F, Ellis J, Clark EG, Harding J, Espuna E, Botner A, et al. Novel porcine circoviruses from pigs with wasting disease syndromes. Vet Rec. 1998;142(17):467–468. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

