Genome-Wide Identification and Expression Analysis of the KUP Family under Abiotic Stress in Cassava (Manihot esculenta Crantz)
- PMID: 29416511
- PMCID: PMC5787556
- DOI: 10.3389/fphys.2018.00017
Genome-Wide Identification and Expression Analysis of the KUP Family under Abiotic Stress in Cassava (Manihot esculenta Crantz)
Abstract
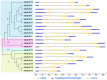
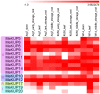
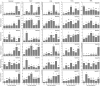
KT/HAK/KUP (KUP) family is responsible for potassium ion (K+) transport, which plays a vital role in the response of plants to abiotic stress by maintaining osmotic balance. However, our understanding of the functions of the KUP family in the drought-resistant crop cassava (Manihot esculenta Crantz) is limited. In the present study, 21 cassava KUP genes (MeKUPs) were identified and classified into four clusters based on phylogenetic relationships, conserved motifs, and gene structure analyses. Transcriptome analysis revealed the expression diversity of cassava KUPs in various tissues of three genotypes. Comparative transcriptome analysis showed that the activation of MeKUP genes by drought was more in roots than that in leaves of Arg7 and W14 genotypes, whereas less in roots than that in leaves of SC124 variety. These findings indicate that different cassava genotypes utilize various drought resistance mechanism mediated by KUP genes. Specific KUP genes showed broad upregulation after exposure to salt, osmotic, cold, H2O2, and abscisic acid (ABA) treatments. Taken together, this study provides insights into the KUP-mediated drought response of cassava at transcription levels and identifies candidate genes that may be utilized in improving crop tolerance to abiotic stress.
Keywords: KUP family; cassava; drought stress; gene expression; identification.
Figures



Similar articles
-
The ERF transcription factor family in cassava: genome-wide characterization and expression analyses against drought stress.Sci Rep. 2016 Nov 21;6:37379. doi: 10.1038/srep37379. Sci Rep. 2016. PMID: 27869212 Free PMC article.
-
The Late Embryogenesis Abundant Protein Family in Cassava (Manihot esculenta Crantz): Genome-Wide Characterization and Expression during Abiotic Stress.Molecules. 2018 May 17;23(5):1196. doi: 10.3390/molecules23051196. Molecules. 2018. PMID: 29772750 Free PMC article.
-
Involvement of abscisic acid-responsive element-binding factors in cassava (Manihot esculenta) dehydration stress response.Sci Rep. 2019 Sep 2;9(1):12661. doi: 10.1038/s41598-019-49083-3. Sci Rep. 2019. PMID: 31477771 Free PMC article.
-
Plant HAK/KUP/KT K+ transporters: Function and regulation.Semin Cell Dev Biol. 2018 Feb;74:133-141. doi: 10.1016/j.semcdb.2017.07.009. Epub 2017 Jul 13. Semin Cell Dev Biol. 2018. PMID: 28711523 Review.
-
Cassava biology and physiology.Plant Mol Biol. 2004 Nov;56(4):481-501. doi: 10.1007/s11103-005-2270-7. Plant Mol Biol. 2004. PMID: 15669146 Review.
Cited by
-
Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review.Plants (Basel). 2019 Jan 30;8(2):34. doi: 10.3390/plants8020034. Plants (Basel). 2019. PMID: 30704089 Free PMC article. Review.
-
Genome-wide identification, expression profiling, and functional analysis of ammonium transporter 2 (AMT2) gene family in cassava (Manihot esculenta crantz).Front Genet. 2023 Feb 22;14:1145735. doi: 10.3389/fgene.2023.1145735. eCollection 2023. Front Genet. 2023. PMID: 36911399 Free PMC article.
-
The Sweet Potato K+ Transporter IbHAK11 Regulates K+ Deficiency and High Salinity Stress Tolerance by Maintaining Positive Ion Homeostasis.Plants (Basel). 2023 Jun 23;12(13):2422. doi: 10.3390/plants12132422. Plants (Basel). 2023. PMID: 37446983 Free PMC article.
-
Genome-wide characterization and expression analysis of the HAK gene family in response to abiotic stresses in Medicago.BMC Genomics. 2022 Dec 1;23(1):791. doi: 10.1186/s12864-022-09009-2. BMC Genomics. 2022. PMID: 36456911 Free PMC article.
-
Molecular Evolution and Expansion of the KUP Family in the Allopolyploid Cotton Species Gossypium hirsutum and Gossypium barbadense.Front Plant Sci. 2020 Sep 30;11:545042. doi: 10.3389/fpls.2020.545042. eCollection 2020. Front Plant Sci. 2020. PMID: 33101325 Free PMC article.
References
-
- Amrutha R. N., Sekhar P. N., Varshney R. K., Kishor P. B. K. (2007). Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci. 172, 708–721. 10.1016/j.plantsci.2006.11.019 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

