Disruption of zinc finger DNA binding domain in catabolite repressor Mig1 increases growth rate, hyphal branching, and cellulase expression in hypercellulolytic fungus Penicillium funiculosum NCIM1228
- PMID: 29416560
- PMCID: PMC5784589
- DOI: 10.1186/s13068-018-1011-5
Disruption of zinc finger DNA binding domain in catabolite repressor Mig1 increases growth rate, hyphal branching, and cellulase expression in hypercellulolytic fungus Penicillium funiculosum NCIM1228
Abstract
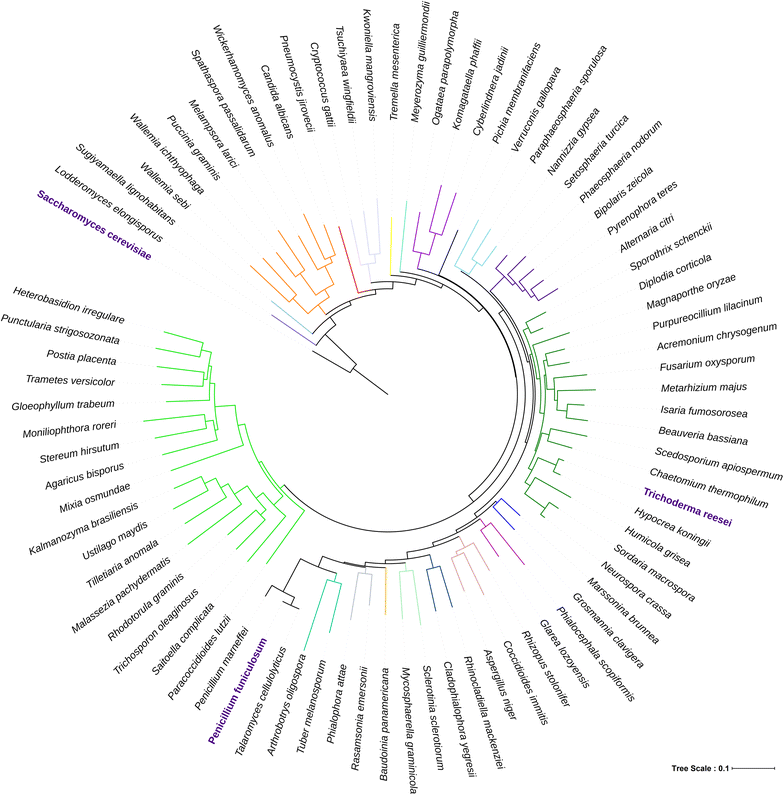
Background: There is an urgent requirement for second-generation bio-based industries for economical yet efficient enzymatic cocktail to convert diverse cellulosic biomass into fermentable sugars. In our previous study, secretome of Penicillium funiculosum NCIM1228 showed high commercial potential by exhibiting high biomass hydrolyzing efficiency. To develop NCIM1228 further as an industrial workhorse, one of the major genetic interventions needed is global deregulation of cellulolytic genes to achieve higher enzyme production. Mig1 orthologs found in all yeast and filamentous fungi are transcriptional regulators that maintain carbon homeostasis by negatively regulating genes of secondary carbon source utilization. Their disruption has long been known to be beneficial for increasing the production of secreted enzymes for alternate carbon source utilization.
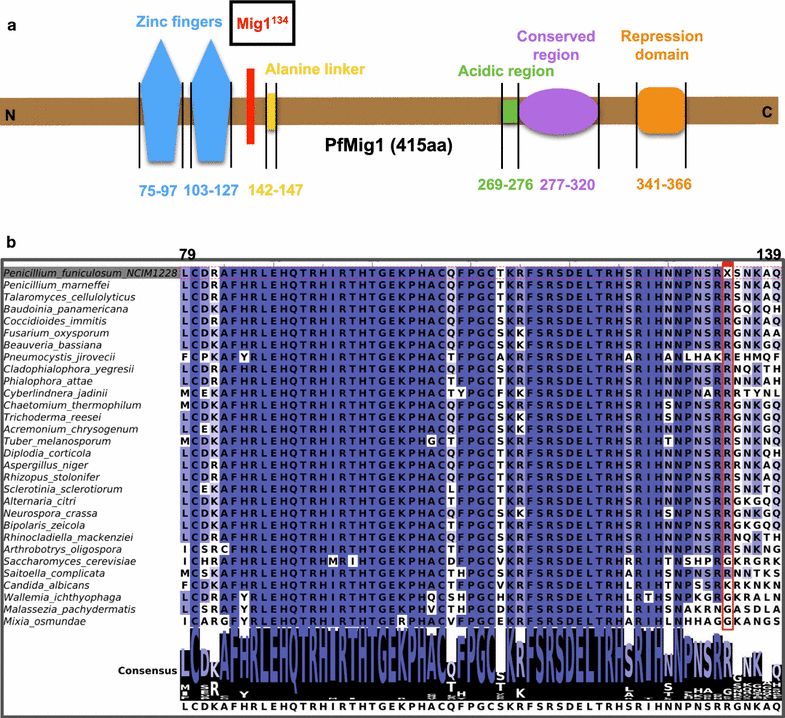
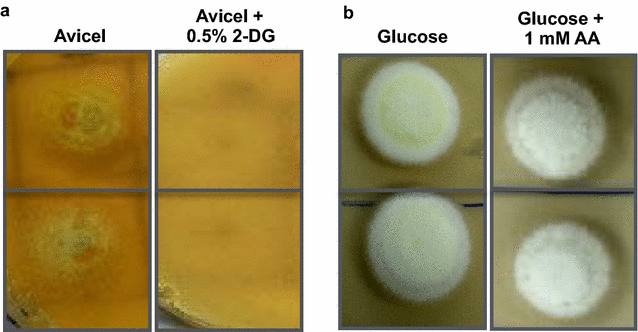
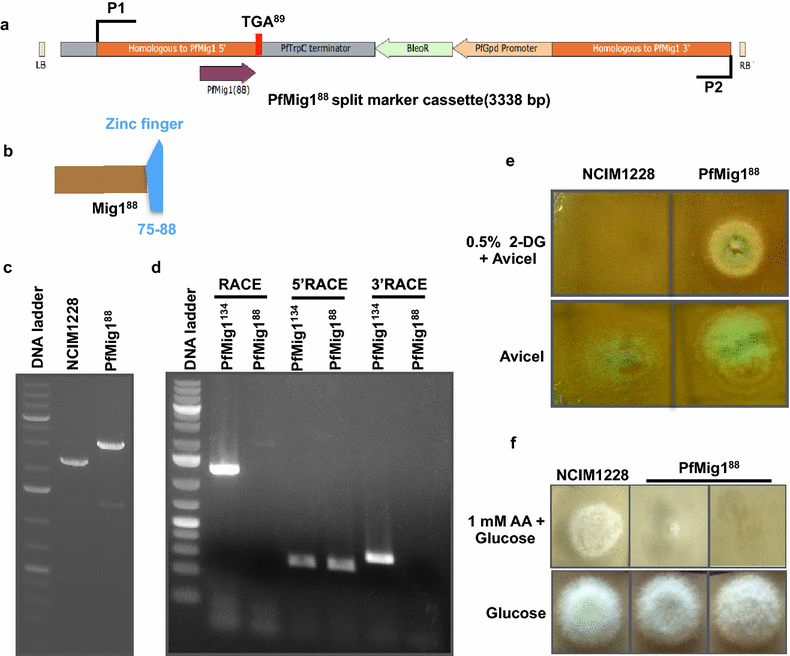
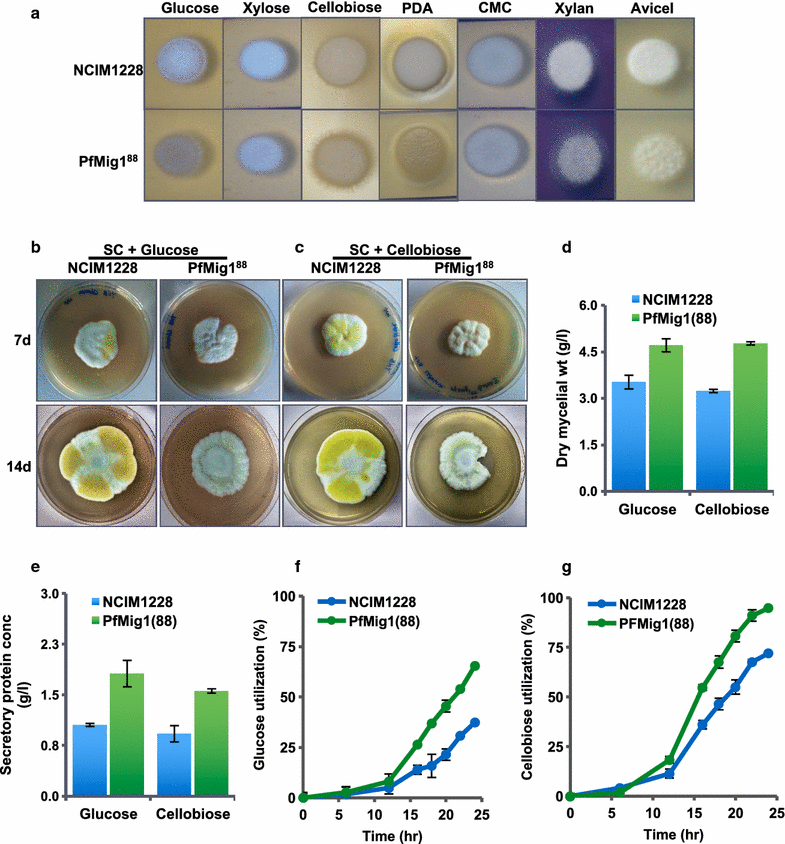
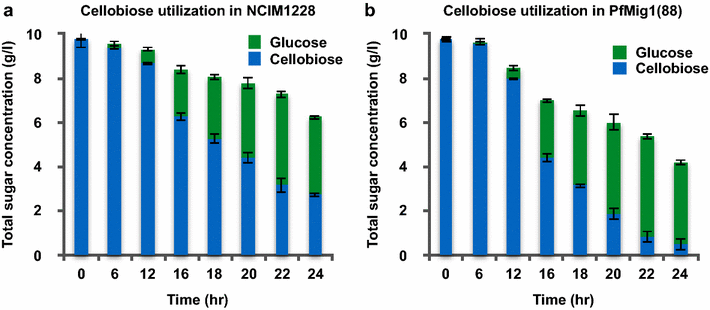
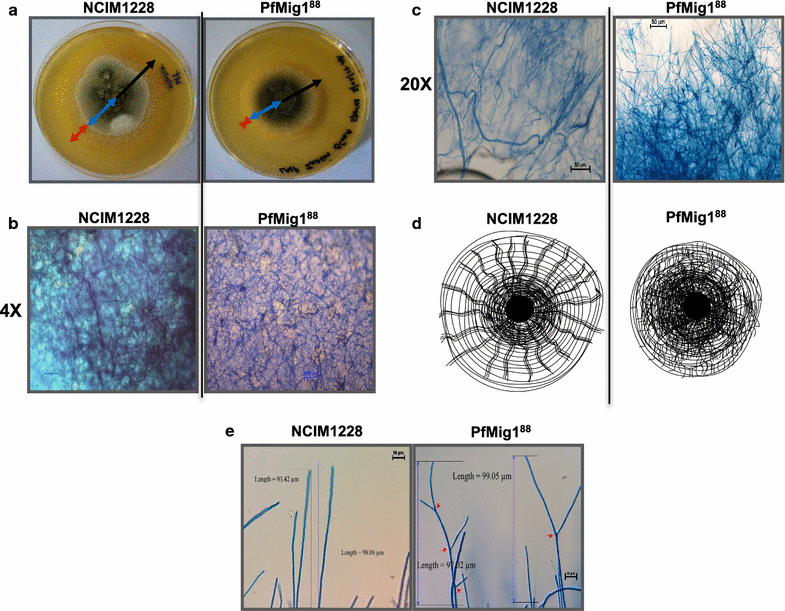
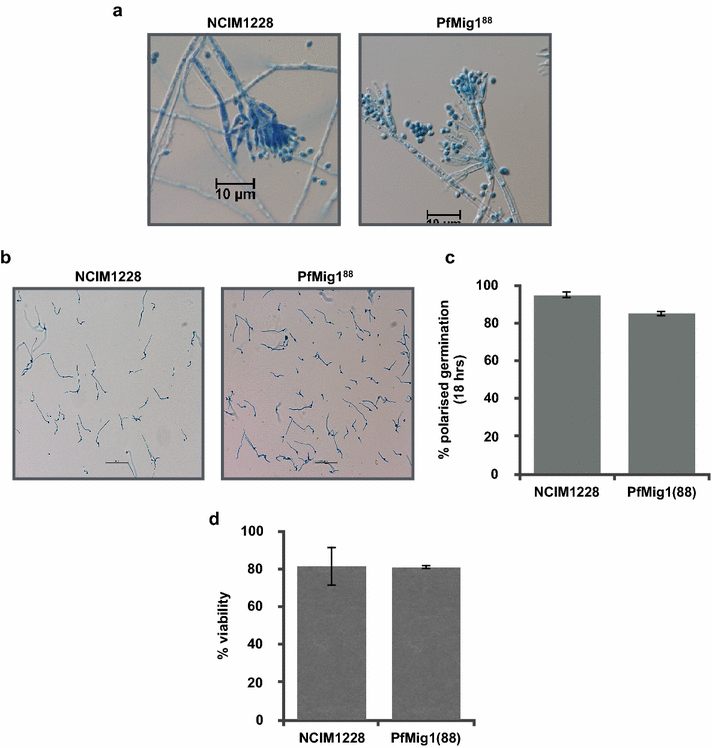
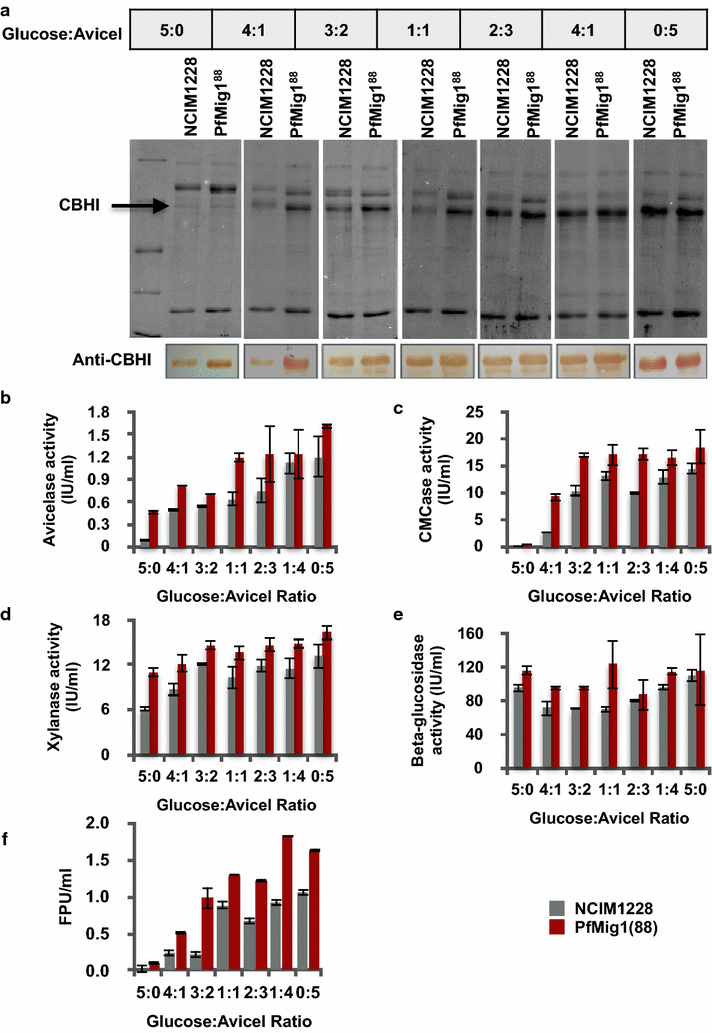
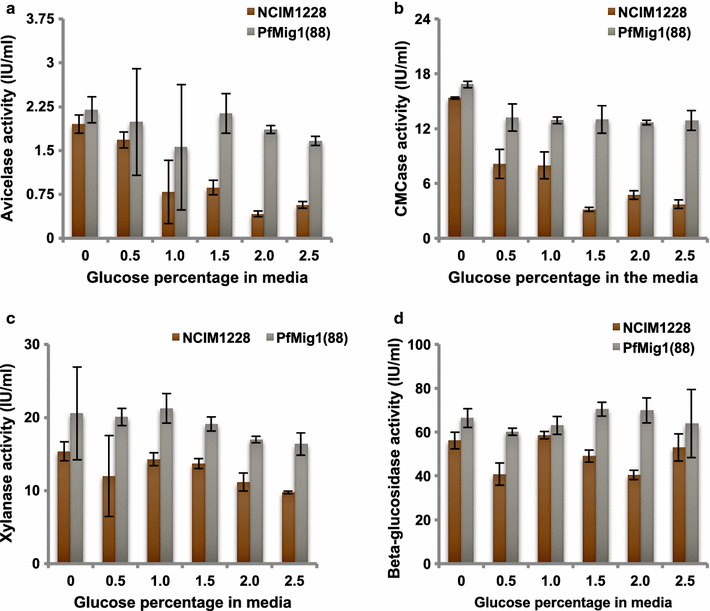
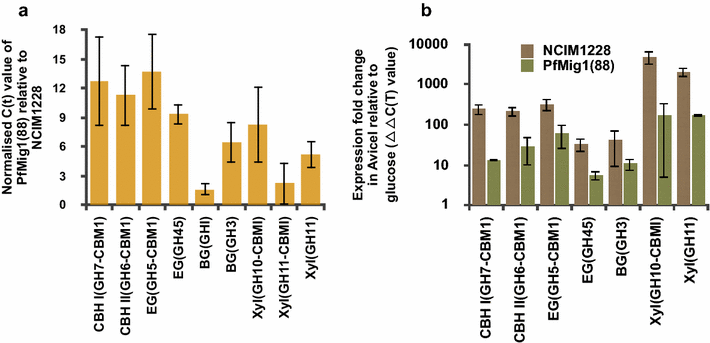
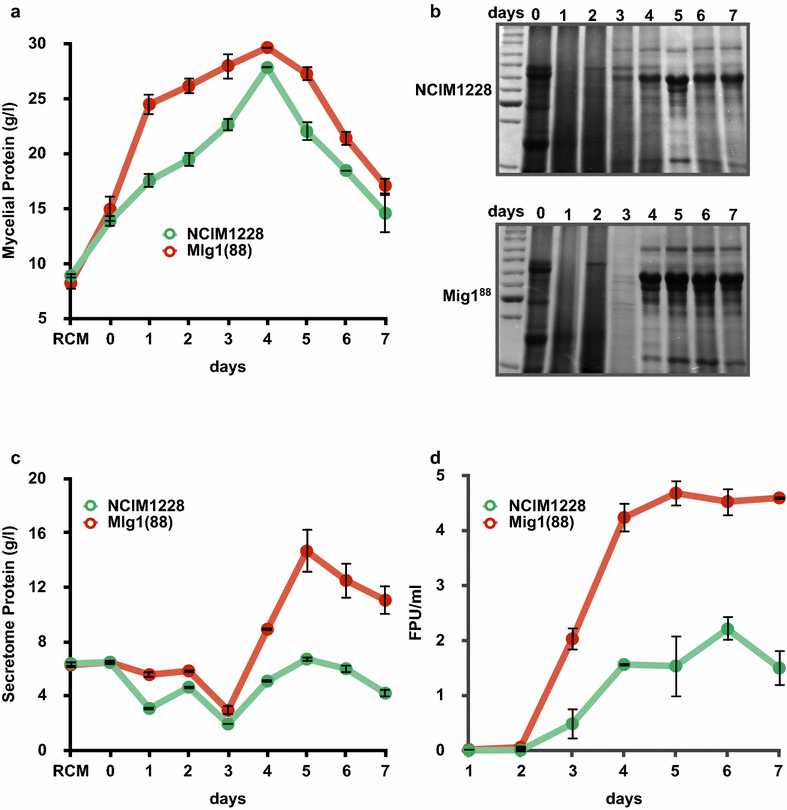
Results: Upon detailed genotypic and phenotypic analysis, we observed that NCIM1228 harbors a truncated yet functional allele of homolog of a well-known catabolite repressor, Mig1. Alleviation of carbon repression in NCIM1228 was attained by replacing functional Mig1134 allele with null allele Mig188. P. funiculosum having Mig188 null allele showed better growth characteristics and 1.75-fold better glucose utilization than parent strain. We also showed that visibly small colony size, one of the major characteristics of CCR disruptant strains in filamentous fungi, was not due to retarded growth, but altered hyphal morphology. CCR-disrupted strain PfMig188 showed profuse branching pattern in terminal hyphae resulting in small and compact colonies with compromised filamentous proliferation. We further observed that basal level expression of two major classes of cellulases, namely, cellobiohydrolase and endoglucanase, was regulated by Mig1134 in NCIM1228, whereas other two major classes, namely, xylanases and β-glucosidase, were only marginally regulated. Finally, CCR disruption in P. funiculosum NCIM1228 led to prolonged cellulase induction in production medium resulting in twofold increased cellulase activity than the parent strain with maximum secreted protein titer being > 14 g/l.
Conclusions: CCR-disrupted P. funiculosum showed better growth, enhanced carbon source utilization, profuse branching pattern in terminal hyphae, and higher cellulase activity than parent strain. Our findings are particularly important in shedding light on important functions performed by Mig1 in addition to its role as negative regulator of alternate carbon source utilization in filamentous fungi.
Keywords: Carbon catabolite repression; Cellulases; De-repression; Mig1 orthologs; Zinc finger domains.
Figures
References
-
- Ranjita Biswas APAVSB . Production of cellulolytic enzymes. New York: Wiley; 2014.
-
- Sajith SPP, Sreedevi S, Benjamin S. An overview on fungal cellulases with an industrial perspective. J Nutr Food Sci. 2016;6:1.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

