The dynamic intein landscape of eukaryotes
- PMID: 29416568
- PMCID: PMC5784728
- DOI: 10.1186/s13100-018-0111-x
The dynamic intein landscape of eukaryotes
Abstract
Background: Inteins are mobile, self-splicing sequences that interrupt proteins and occur across all three domains of life. Scrutiny of the intein landscape in prokaryotes led to the hypothesis that some inteins are functionally important. Our focus shifts to eukaryotic inteins to assess their diversity, distribution, and dissemination, with the aim to comprehensively evaluate the eukaryotic intein landscape, understand intein maintenance, and dissect evolutionary relationships.
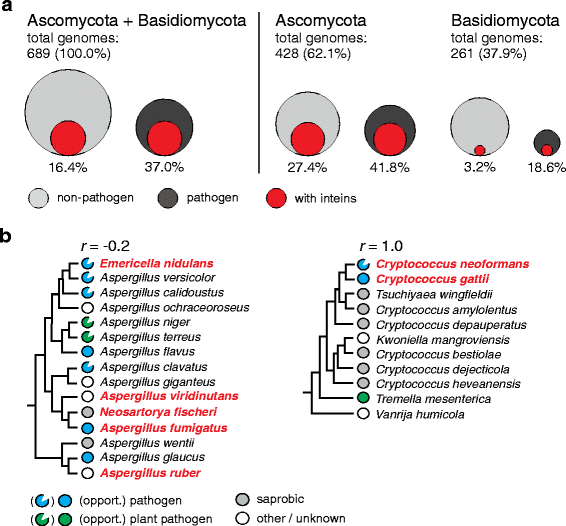
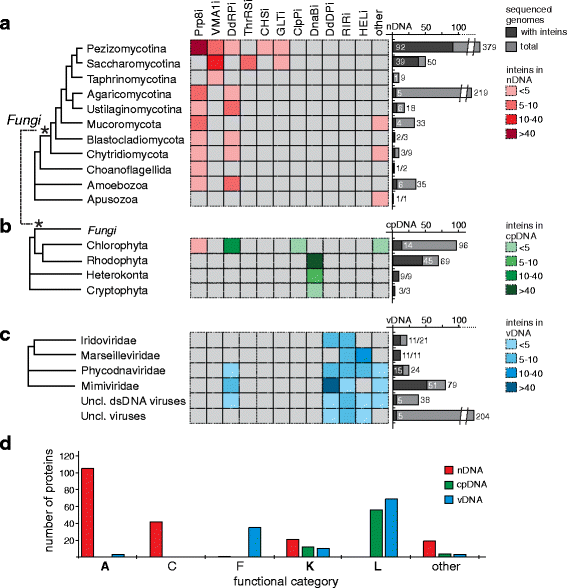
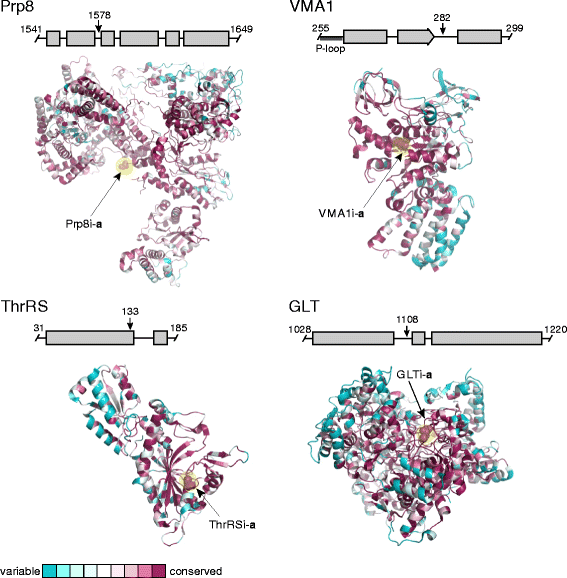
Results: This bioinformatics study reveals that eukaryotic inteins are scarce, but present in nuclear genomes of fungi, chloroplast genomes of algae, and within some eukaryotic viruses. There is a preponderance of inteins in several fungal pathogens of humans and plants. Inteins are pervasive in certain proteins, including the nuclear RNA splicing factor, Prp8, and the chloroplast DNA helicase, DnaB. We find that eukaryotic inteins frequently localize to unstructured loops of the host protein, often at highly conserved sites. More broadly, a sequence similarity network analysis of all eukaryotic inteins uncovered several routes of intein mobility. Some eukaryotic inteins appear to have been acquired through horizontal transfer with dsDNA viruses, yet other inteins are spread through intragenomic transfer. Remarkably, endosymbiosis can explain patterns of DnaB intein inheritance across several algal phyla, a novel mechanism for intein acquisition and distribution.
Conclusions: Overall, an intriguing picture emerges for how the eukaryotic intein landscape arose, with many evolutionary forces having contributed to its current state. Our collective results provide a framework for exploring inteins as novel regulatory elements and innovative drug targets.
Keywords: Endosymbiosis; Horizontal transfer; Intein; Mobile elements; Sequence similarity network.
Conflict of interest statement
Not applicable.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

