Mitochondrial dysfunction activates lysosomal-dependent mitophagy selectively in cancer cells
- PMID: 29416672
- PMCID: PMC5787530
- DOI: 10.18632/oncotarget.23171
Mitochondrial dysfunction activates lysosomal-dependent mitophagy selectively in cancer cells
Abstract
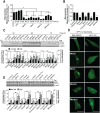
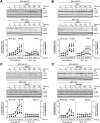
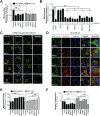
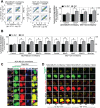
Molecules designed to target and accumulate in the mitochondria are an emerging therapeutic approach for cancer and other indications. Mitochondria-targeted redox agents (MTAs) induce mitochondrial damage and autophagy in cancer cells. However, the mechanisms for these molecules to induce mitophagy, the clearance of damaged mitochondria, are largely unknown. Using breast derived cell lines and a series of targeted molecules, mitochondrial dysfunction and autophagy was established to be selective for MDA-MB-231 cancer cells as compared to the non-cancerous MCF-12A cells. Kinetic analyses revealed that mitochondrial dysfunction precedes the activation of autophagy in these cancer cells. To determine the onset of mitophagy, stably expressing mitochondrial mKeima, a mitochondrial pH sensor, cell lines were generated and revealed that these drugs activate lysosomal dependent mitochondrial degradation in MDA-MB-231 cells. Mitophagy was confirmed by identifying the accumulation of a PINK1, mitochondria located in autophagosomes, and the formation of an autophagosome-mitochondria protein (MFN2-LC3-II) complex. These results are the first to demonstrate that mitochondrial redox agents selectively induce mitophagy in a breast cancer cell line and their potential application both as tools for investigating mitochondrial biomechanics and as therapeutic strategies that target mitochondrial metabolism.
Keywords: autophagy; cancer; mitochondria; mitophagy; mitoquinone.
Conflict of interest statement
CONFLICTS OF INTEREST The authors have no conflicts of interests. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the United States Food and Drug Administration and the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government.
Figures


References
-
- Rao VA, Klein SR, Bonar SJ, Zielonka J, Mizuno N, Dickey JS, Keller PW, Joseph J, Kalyanaraman B, Shacter E. The antioxidant transcription factor Nrf2 negatively regulates autophagy and growth arrest induced by the anticancer redox agent mitoquinone. J Biol Chem. 2010;285:34447–59. https://doi.org/10.1074/jbc.M110.133579. - DOI - PMC - PubMed
-
- Cheng G, Zielonka J, Ouari O, Lopez M, McAllister D, Boyle K, Barrios CS, Weber JJ, Johnson BD, Hardy M, Dwinell MB, Kalyanaraman B. Mitochondria-targeted analogues of metformin exhibit enhanced antiproliferative and radiosensitizing effects in pancreatic cancer cells. Cancer Res. 2016;76:3904–15. https://doi.org/10.1158/0008-5472.CAN-15-2534. - DOI - PMC - PubMed
-
- Murphy MP. Targeting lipophilic cations to mitochondria. Biochim Biophys Acta. 2008;1777:1028–31. https://doi.org/10.1016/j.bbabio.2008.03.029. - DOI - PubMed
-
- Smith RA, Adlam VJ, Blaikie FH, Manas AR, Porteous CM, James AM, Ross MF, Logan A, Cocheme HM, Trnka J, Prime TA, Abakumova I, Jones BA, et al. Mitochondria-targeted antioxidants in the treatment of disease. Ann N Y Acad Sci. 2008;1147:105–11. https://doi.org/10.1196/annals.1427.003. - DOI - PubMed
-
- Wen S, Zhu D, Huang P. Targeting cancer cell mitochondria as a therapeutic approach. Future Med Chem. 2013;5:53–67. https://doi.org/10.4155/fmc.12.190. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

