A 250 plastome phylogeny of the grass family (Poaceae): topological support under different data partitions
- PMID: 29416954
- PMCID: PMC5798404
- DOI: 10.7717/peerj.4299
A 250 plastome phylogeny of the grass family (Poaceae): topological support under different data partitions
Abstract
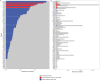
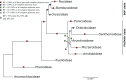
The systematics of grasses has advanced through applications of plastome phylogenomics, although studies have been largely limited to subfamilies or other subgroups of Poaceae. Here we present a plastome phylogenomic analysis of 250 complete plastomes (179 genera) sampled from 44 of the 52 tribes of Poaceae. Plastome sequences were determined from high throughput sequencing libraries and the assemblies represent over 28.7 Mbases of sequence data. Phylogenetic signal was characterized in 14 partitions, including (1) complete plastomes; (2) protein coding regions; (3) noncoding regions; and (4) three loci commonly used in single and multi-gene studies of grasses. Each of the four main partitions was further refined, alternatively including or excluding positively selected codons and also the gaps introduced by the alignment. All 76 protein coding plastome loci were found to be predominantly under purifying selection, but specific codons were found to be under positive selection in 65 loci. The loci that have been widely used in multi-gene phylogenetic studies had among the highest proportions of positively selected codons, suggesting caution in the interpretation of these earlier results. Plastome phylogenomic analyses confirmed the backbone topology for Poaceae with maximum bootstrap support (BP). Among the 14 analyses, 82 clades out of 309 resolved were maximally supported in all trees. Analyses of newly sequenced plastomes were in agreement with current classifications. Five of seven partitions in which alignment gaps were removed retrieved Panicoideae as sister to the remaining PACMAD subfamilies. Alternative topologies were recovered in trees from partitions that included alignment gaps. This suggests that ambiguities in aligning these uncertain regions might introduce a false signal. Resolution of these and other critical branch points in the phylogeny of Poaceae will help to better understand the selective forces that drove the radiation of the BOP and PACMAD clades comprising more than 99.9% of grass diversity.
Keywords: Aristidoideae; Classification; Grasses; Incongruence; Panicoideae; Phylogenomics; Plastome; Positive selection; Purifying selection; Systematics.
Conflict of interest statement
Joseph M. Craine is an employee of Jonah Ventures and an Academic Editor for PeerJ.
Figures






References
-
- Acosta JM, Scataglini MA, Reinheimer R, Zuloaga FO. A phylogenetic study of subtribe Otachyriinae (Poaceae, Panicoideae, Paspaleae) Plant Systematics and Evolution. 2014;300(10):2155–2166. doi: 10.1007/s00606-014-1034-8. - DOI
-
- Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19(6):716–723. doi: 10.1109/TAC.1974.1100705. - DOI
-
- Attigala L, Triplett JK, Kathriarachchi H-S, Clark LG. A new genus and a major temperate bamboo lineage of the Arundinarieae (Poaceae: Bambusoideae) from Sri Lanka based on a multi-locus plastid phylogeny. Phytotaxa. 2014;174(4):187–205. doi: 10.11646/phytotaxa.174.4.1. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

