MELK expression correlates with tumor mitotic activity but is not required for cancer growth
- PMID: 29417930
- PMCID: PMC5805410
- DOI: 10.7554/eLife.32838
MELK expression correlates with tumor mitotic activity but is not required for cancer growth
Abstract
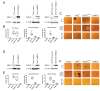
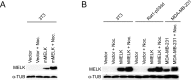
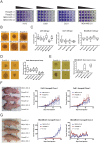
The Maternal Embryonic Leucine Zipper Kinase (MELK) has been identified as a promising therapeutic target in multiple cancer types. MELK over-expression is associated with aggressive disease, and MELK has been implicated in numerous cancer-related processes, including chemotherapy resistance, stem cell renewal, and tumor growth. Previously, we established that triple-negative breast cancer cell lines harboring CRISPR/Cas9-induced null mutations in MELK proliferate at wild-type levels in vitro (<xref ref-type="bibr" rid="bib34">Lin et al., 2017</xref>). Here, we generate several additional knockout clones of MELK and demonstrate that across cancer types, cells lacking MELK exhibit wild-type growth in vitro, under environmental stress, in the presence of cytotoxic chemotherapies, and in vivo. By combining our MELK-knockout clones with a recently described, highly specific MELK inhibitor, we further demonstrate that the acute inhibition of MELK results in no specific anti-proliferative phenotype. Analysis of gene expression data from cohorts of cancer patients identifies MELK expression as a correlate of tumor mitotic activity, explaining its association with poor clinical prognosis. In total, our results demonstrate the power of CRISPR/Cas9-based genetic approaches to investigate cancer drug targets, and call into question the rationale for treating patients with anti-MELK monotherapies.
Keywords: CRISPR/Cas9; biomarkers; cancer biology; cell cycle; drug targets; human; mitotic kinase.
© 2018, Giuliano et al.
Conflict of interest statement
CG, AL, AP, JS No competing interests declared, JS Joan C. Smith is affiliated with Google Inc. The author has no financial interests to declare.
Figures






Comment in
- doi: 10.7554/eLife.32402
References
-
- Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, Fong S, van Lookeren Campagne M, Godowski P, Williams PM, Chan AC, Clark HF. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes & Immunity. 2005;6:319–331. doi: 10.1038/sj.gene.6364173. - DOI - PubMed
-
- Beke L, Kig C, Linders JT, Boens S, Boeckx A, van Heerde E, Parade M, De Bondt A, Van den Wyngaert I, Bashir T, Ogata S, Meerpoel L, Van Eynde A, Johnson CN, Beullens M, Brehmer D, Bollen M. MELK-T1, a small-molecule inhibitor of protein kinase MELK, decreases DNA-damage tolerance in proliferating cancer cells. Bioscience Reports. 2015;35:e00267. doi: 10.1042/BSR20150194. - DOI - PMC - PubMed
-
- Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Research. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

