Evolutionary conservation of the allosteric activation of factor VIIa by tissue factor in lamprey
- PMID: 29418058
- PMCID: PMC5893411
- DOI: 10.1111/jth.13968
Evolutionary conservation of the allosteric activation of factor VIIa by tissue factor in lamprey
Abstract
Essentials Tissue factor (TF) enhances factor VIIa (FVIIa) activity through structural and dynamic changes. We analyzed conservation of TF-activated FVIIa allosteric networks in extant vertebrate lamprey. Lamprey Tf/FVIIa molecular dynamics show conserved Tf-induced structural/dynamic FVIIa changes. Lamprey Tf activation of FVIIa allosteric networks follows molecular pathways similar to human.
Summary: Background Previous studies have provided insight into the molecular basis of human tissue factor (TF) activation of activated factor VII (FVIIa). TF-induced allosteric networks of FVIIa activation have been rationalized through analysis of the dynamic changes and residue connectivities in the human soluble TF (sTF)/FVIIa complex structure during molecular dynamics (MD) simulation. Evolutionary conservation of the molecular mechanisms for TF-induced allosteric FVIIa activation between humans and extant vertebrate jawless fish (lampreys), where blood coagulation emerged more than 500 million years ago, is unknown and of considerable interest. Objective To model the sTf/FVIIa complex from cloned Petromyzon marinus lamprey sequences, and with comparisons to human sTF/FVlla investigate conservation of allosteric mechanisms of FVIIa activity enhancement by soluble TF using MD simulations. Methods Full-length cDNAs of lamprey tf and f7 were cloned and characterized. Comparative models of lamprey sTf/FVIIa complex and free FVIIa were determined based on constructed human sTF/FVIIa complex and free FVIIa models, used in full-atomic MD simulations, and characterized using dynamic network analysis approaches. Results Allosteric paths of correlated motion from Tf contact points in lamprey sTf/FVIIa to the FVIIa active site were determined and quantified, and were found to encompass residue-residue interactions along significantly similar paths compared with human. Conclusions Despite low conservation of residues between lamprey and human proteins, 30% TF and 39% FVII, the structural and protein dynamic effects of TF activation of FVIIa appear conserved and, moreover, present in extant vertebrate proteins from 500 million years ago when TF/FVIIa-initiated extrinsic pathway blood coagulation emerged.
Keywords: allosteric regulation; blood coagulation factors; factor VIIa; thromboplastin; tissue factor.
© 2018 International Society on Thrombosis and Haemostasis.
Conflict of interest statement
The authors state that they have no conflict of interest.
Figures


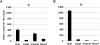


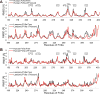
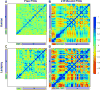

Comment in
-
Evolutionary conservation of the allosteric activation of factor VIIa by tissue factor in lamprey: comment.J Thromb Haemost. 2018 Jul;16(7):1450-1454. doi: 10.1111/jth.14142. Epub 2018 Jun 6. J Thromb Haemost. 2018. PMID: 29733494 No abstract available.
-
Evolutionary conservation of the allosteric activation of factor VIIa by tissue factor in lamprey: reply.J Thromb Haemost. 2018 Jul;16(7):1454-1456. doi: 10.1111/jth.14141. Epub 2018 Jun 6. J Thromb Haemost. 2018. PMID: 29734527 No abstract available.
References
-
- Davidson CJ, Tuddenham EG, McVey JH. 450 million years of hemostasis. J Thromb Haemost. 2003;1:1487–94. - PubMed
-
- Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry. 1991;30:10363–70. - PubMed
-
- Chung DW, Xu W, Davie EW. The blood coagulation factors and inhibitors: their primary structure, complementary DNAs, genes, and expression. In: Marder VJ, Aird WC, Bennet JS, Schulman S, White GC, editors. Hemostasis and Thrombosis, Basic Principles and Clinical Practice. 6th. Philadelphia: Wolters Kluwer, Lippincott Williams & Wilkins; 2013. pp. 110–45.
-
- Morrissey JH, Broze GJ. Tissue factor and the initiation and regulation (TFPI) of coagulation. In: Marder VJ, Aird WC, Bennett JS, Schulman S, White GC, editors. Hemostasis and Thrombosis: Basic Principles and Clinical Practice. 6th. Philadelphia: Lippincott Williams & Wilkins; 2013.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

