Intrinsic Properties of tRNA Molecules as Deciphered via Bayesian Network and Distribution Divergence Analysis
- PMID: 29419741
- PMCID: PMC5871937
- DOI: 10.3390/life8010005
Intrinsic Properties of tRNA Molecules as Deciphered via Bayesian Network and Distribution Divergence Analysis
Abstract
The identity/recognition of tRNAs, in the context of aminoacyl tRNA synthetases (and other molecules), is a complex phenomenon that has major implications ranging from the origins and evolution of translation machinery and genetic code to the evolution and speciation of tRNAs themselves to human mitochondrial diseases to artificial genetic code engineering. Deciphering it via laboratory experiments, however, is difficult and necessarily time- and resource-consuming. In this study, we propose a mathematically rigorous two-pronged in silico approach to identifying and classifying tRNA positions important for tRNA identity/recognition, rooted in machine learning and information-theoretic methodology. We apply Bayesian Network modeling to elucidate the structure of intra-tRNA-molecule relationships, and distribution divergence analysis to identify meaningful inter-molecule differences between various tRNA subclasses. We illustrate the complementary application of these two approaches using tRNA examples across the three domains of life, and identify and discuss important (informative) positions therein. In summary, we deliver to the tRNA research community a novel, comprehensive methodology for identifying the specific elements of interest in various tRNA molecules, which can be followed up by the corresponding experimental work and/or high-resolution position-specific statistical analyses.
Keywords: bayesian networks; distribution divergence; information theory; operational code; tRNA identity; tRNA recognition.
Conflict of interest statement
The authors declare no conflict of interest.The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures

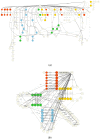
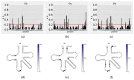
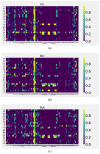
References
-
- Giegé R., Eriani G. eLS. John Wiley & Sons, Ltd.; Hoboken, NJ, USA: 2014. Transfer RNA Recognition and Aminoacylation by Synthetases.
LinkOut - more resources
Full Text Sources
Other Literature Sources

