Live Dynamics of 53BP1 Foci Following Simultaneous Induction of Clustered and Dispersed DNA Damage in U2OS Cells
- PMID: 29419809
- PMCID: PMC5855741
- DOI: 10.3390/ijms19020519
Live Dynamics of 53BP1 Foci Following Simultaneous Induction of Clustered and Dispersed DNA Damage in U2OS Cells
Abstract
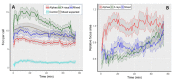
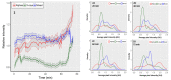
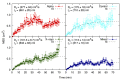
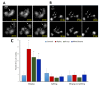
Cells react differently to clustered and dispersed DNA double strand breaks (DSB). Little is known about the initial reaction to simultaneous induction of DSBs with different complexities. Here, we used live cell microscopy to analyse the behaviour of 53BP1-GFP (green fluorescence protein) foci formation at DSBs induced in U2OS cells by alpha particles, X-rays or mixed beams over a 75 min period post irradiation. X-ray-induced foci rapidly increased and declined over the observation interval. After an initial increase, mixed beam-induced foci remained at a constant level over the observation interval, similarly as alpha-induced foci. The average areas of radiation-induced foci were similar for mixed beams and X-rays, being significantly smaller than those induced by alpha particles. Pixel intensities were highest for mixed beam-induced foci and showed the lowest level of variability over time as compared to foci induced by alphas and X-rays alone. Finally, mixed beam-exposed foci showed the lowest level of mobility as compared to alpha and X-ray exposure. The results suggest paralysation of chromatin around foci containing clustered DNA damage.
Keywords: 53BP1 foci; X-rays; alpha particles; clustered DSB; live cell imaging.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., III, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

