Selective Photoaffinity Probe That Enables Assessment of Cannabinoid CB2 Receptor Expression and Ligand Engagement in Human Cells
- PMID: 29420021
- PMCID: PMC5958339
- DOI: 10.1021/jacs.7b11281
Selective Photoaffinity Probe That Enables Assessment of Cannabinoid CB2 Receptor Expression and Ligand Engagement in Human Cells
Abstract
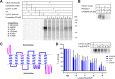
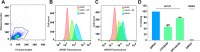
Chemical tools and methods that report on G protein-coupled receptor (GPCR) expression levels and receptor occupancy by small molecules are highly desirable. We report the development of LEI121 as a photoreactive probe to study the type 2 cannabinoid receptor (CB2R), a promising GPCR to treat tissue injury and inflammatory diseases. LEI121 is the first CB2R-selective bifunctional probe that covalently captures CB2R upon photoactivation. An incorporated alkyne serves as ligation handle for the introduction of reporter groups. LEI121 enables target engagement studies and visualization of endogenously expressed CB2R in HL-60 as well as primary human immune cells using flow cytometry. Our findings show that strategically functionalized probes allow monitoring of endogenous GPCR expression and engagement in human cells using tandem photoclick chemistry and hold promise as biomarkers in translational drug discovery.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Discovery of a Photoaffinity Probe that Captures the Active Conformation of the Cannabinoid CB2 Receptor.Chembiochem. 2024 Apr 2;25(7):e202300785. doi: 10.1002/cbic.202300785. Epub 2024 Mar 1. Chembiochem. 2024. PMID: 38372466
-
Novel pyridine derivatives as potent and selective CB2 cannabinoid receptor agonists.Bioorg Med Chem Lett. 2009 Oct 15;19(20):5931-5. doi: 10.1016/j.bmcl.2009.08.063. Epub 2009 Aug 21. Bioorg Med Chem Lett. 2009. PMID: 19736007
-
Structure-kinetic relationship studies of cannabinoid CB2 receptor agonists reveal substituent-specific lipophilic effects on residence time.Biochem Pharmacol. 2018 Jun;152:129-142. doi: 10.1016/j.bcp.2018.03.018. Epub 2018 Mar 21. Biochem Pharmacol. 2018. PMID: 29574067
-
Targeting the cannabinoid CB2 receptor: mutations, modeling and development of CB2 selective ligands.Curr Med Chem. 2005;12(10):1217-37. doi: 10.2174/0929867053764617. Curr Med Chem. 2005. PMID: 15892633 Review.
-
Cannabinoids in pain management: CB1, CB2 and non-classic receptor ligands.Expert Opin Investig Drugs. 2014 Aug;23(8):1123-40. doi: 10.1517/13543784.2014.918603. Epub 2014 May 16. Expert Opin Investig Drugs. 2014. PMID: 24836296 Review.
Cited by
-
A fluorescent photoaffinity probe for formyl peptide receptor 1 labelling in living cells.RSC Chem Biol. 2023 Jan 13;4(3):216-222. doi: 10.1039/d2cb00199c. eCollection 2023 Mar 8. RSC Chem Biol. 2023. PMID: 36908701 Free PMC article.
-
Chemical Proteomics Reveals Off-Targets of the Anandamide Reuptake Inhibitor WOBE437.ACS Chem Biol. 2022 May 20;17(5):1174-1183. doi: 10.1021/acschembio.2c00122. Epub 2022 Apr 28. ACS Chem Biol. 2022. PMID: 35482948 Free PMC article.
-
Development of Covalent Ligands for G Protein-Coupled Receptors: A Case for the Human Adenosine A3 Receptor.J Med Chem. 2019 Apr 11;62(7):3539-3552. doi: 10.1021/acs.jmedchem.8b02026. Epub 2019 Mar 28. J Med Chem. 2019. PMID: 30869893 Free PMC article.
-
Development of a Cannabinoid-Based Photoaffinity Probe to Determine the Δ8/9-Tetrahydrocannabinol Protein Interaction Landscape in Neuroblastoma Cells.Cannabis Cannabinoid Res. 2018 Jul 1;3(1):136-151. doi: 10.1089/can.2018.0003. eCollection 2018. Cannabis Cannabinoid Res. 2018. PMID: 29992186 Free PMC article.
-
Unbiased Identification of the Liposome Protein Corona using Photoaffinity-based Chemoproteomics.ACS Cent Sci. 2020 Apr 22;6(4):535-545. doi: 10.1021/acscentsci.9b01222. Epub 2020 Apr 1. ACS Cent Sci. 2020. PMID: 32342003 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

