HER kinase inhibition in patients with HER2- and HER3-mutant cancers
- PMID: 29420467
- PMCID: PMC5808581
- DOI: 10.1038/nature25475
HER kinase inhibition in patients with HER2- and HER3-mutant cancers
Erratum in
-
Author Correction: HER kinase inhibition in patients with HER2- and HER3-mutant cancers.Nature. 2019 Feb;566(7745):E11-E12. doi: 10.1038/s41586-019-0974-0. Nature. 2019. PMID: 30755741
Abstract
Somatic mutations of ERBB2 and ERBB3 (which encode HER2 and HER3, respectively) are found in a wide range of cancers. Preclinical modelling suggests that a subset of these mutations lead to constitutive HER2 activation, but most remain biologically uncharacterized. Here we define the biological and therapeutic importance of known oncogenic HER2 and HER3 mutations and variants of unknown biological importance by conducting a multi-histology, genomically selected, 'basket' trial using the pan-HER kinase inhibitor neratinib (SUMMIT; clinicaltrials.gov identifier NCT01953926). Efficacy in HER2-mutant cancers varied as a function of both tumour type and mutant allele to a degree not predicted by preclinical models, with the greatest activity seen in breast, cervical and biliary cancers and with tumours that contain kinase domain missense mutations. This study demonstrates how a molecularly driven clinical trial can be used to refine our biological understanding of both characterized and new genomic alterations with potential broad applicability for advancing the paradigm of genome-driven oncology.
Conflict of interest statement
Competing financial interests include the following: R.E.C., F.X., L.D.E., G.M., C.F., A.S.L., and R.P.B are employees of Puma Biotechnology. D.M.H., M.S., and J.B. receive research support from Puma Biotechnology, B.T.L. and M.S. receive research funding from Diachi, A.D. receives personal fees from Roche, and D.S. received personal fees from Loxo Oncology and Pfizer.
Figures





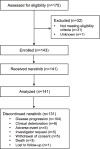
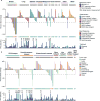

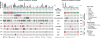
Comment in
-
Many mutations in one clinical-trial basket.Nature. 2018 Feb 8;554(7691):173-175. doi: 10.1038/d41586-018-01312-x. Nature. 2018. PMID: 29420481 No abstract available.
References
-
- Schram A, et al. Landscape of somatic ERBB2 mutations: Findings from AACR GENIE and comparison to ongoing ERBB2 mutant basket study. Cancer Res. 2017;77(13 Supplement):LB-103.
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

