When are pathogen genome sequences informative of transmission events?
- PMID: 29420641
- PMCID: PMC5821398
- DOI: 10.1371/journal.ppat.1006885
When are pathogen genome sequences informative of transmission events?
Abstract
Recent years have seen the development of numerous methodologies for reconstructing transmission trees in infectious disease outbreaks from densely sampled whole genome sequence data. However, a fundamental and as of yet poorly addressed limitation of such approaches is the requirement for genetic diversity to arise on epidemiological timescales. Specifically, the position of infected individuals in a transmission tree can only be resolved by genetic data if mutations have accumulated between the sampled pathogen genomes. To quantify and compare the useful genetic diversity expected from genetic data in different pathogen outbreaks, we introduce here the concept of 'transmission divergence', defined as the number of mutations separating whole genome sequences sampled from transmission pairs. Using parameter values obtained by literature review, we simulate outbreak scenarios alongside sequence evolution using two models described in the literature to describe transmission divergence of ten major outbreak-causing pathogens. We find that while mean values vary significantly between the pathogens considered, their transmission divergence is generally very low, with many outbreaks characterised by large numbers of genetically identical transmission pairs. We describe the impact of transmission divergence on our ability to reconstruct outbreaks using two outbreak reconstruction tools, the R packages outbreaker and phybreak, and demonstrate that, in agreement with previous observations, genetic sequence data of rapidly evolving pathogens such as RNA viruses can provide valuable information on individual transmission events. Conversely, sequence data of pathogens with lower mean transmission divergence, including Streptococcus pneumoniae, Shigella sonnei and Clostridium difficile, provide little to no information about individual transmission events. Our results highlight the informational limitations of genetic sequence data in certain outbreak scenarios, and demonstrate the need to expand the toolkit of outbreak reconstruction tools to integrate other types of epidemiological data.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
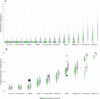

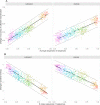
References
-
- Ferguson NM, Donnelly CA, Anderson RM. Transmission intensity and impact of control policies on the foot and mouth epidemic in Great Britain. Nature. 2001;413: 542–548. doi: 10.1038/35097116 - DOI - PubMed
-
- Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am J Epidemiol. 2004;160: 509–516. doi: 10.1093/aje/kwh255 - DOI - PMC - PubMed
-
- Spada E, Sagliocca L, Sourdis J, Garbuglia AR, Poggi V, De Fusco C, et al. Use of the minimum spanning tree model for molecular epidemiological investigation of a nosocomial outbreak of hepatitis C virus infection. J Clin Microbiol. 2004;42: 4230–4236. doi: 10.1128/JCM.42.9.4230-4236.2004 - DOI - PMC - PubMed
-
- Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438: 355–359. doi: 10.1038/nature04153 - DOI - PMC - PubMed
-
- Jombart T, Cori A, Didelot X, Cauchemez S, Fraser C, Ferguson N. Bayesian Reconstruction of Disease Outbreaks by Combining Epidemiologic and Genomic Data. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003457 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

