Open-source chemogenomic data-driven algorithms for predicting drug-target interactions
- PMID: 29420684
- PMCID: PMC6781586
- DOI: 10.1093/bib/bby010
Open-source chemogenomic data-driven algorithms for predicting drug-target interactions
Abstract
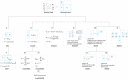
While novel technologies such as high-throughput screening have advanced together with significant investment by pharmaceutical companies during the past decades, the success rate for drug development has not yet been improved prompting researchers looking for new strategies of drug discovery. Drug repositioning is a potential approach to solve this dilemma. However, experimental identification and validation of potential drug targets encoded by the human genome is both costly and time-consuming. Therefore, effective computational approaches have been proposed to facilitate drug repositioning, which have proved to be successful in drug discovery. Doubtlessly, the availability of open-accessible data from basic chemical biology research and the success of human genome sequencing are crucial to develop effective in silico drug repositioning methods allowing the identification of potential targets for existing drugs. In this work, we review several chemogenomic data-driven computational algorithms with source codes publicly accessible for predicting drug-target interactions (DTIs). We organize these algorithms by model properties and model evolutionary relationships. We re-implemented five representative algorithms in R programming language, and compared these algorithms by means of mean percentile ranking, a new recall-based evaluation metric in the DTI prediction research field. We anticipate that this review will be objective and helpful to researchers who would like to further improve existing algorithms or need to choose appropriate algorithms to infer potential DTIs in the projects. The source codes for DTI predictions are available at: https://github.com/minghao2016/chemogenomicAlg4DTIpred.
Keywords: chemogenomic data; drug discovery; drug–target interaction; in silico drug repositioning; mean percentile ranking; open-source code.
Published by Oxford University Press 2018. This work is written by US Government employees and is in the public domain in the US.
Figures



References
-
- Kola I, Landis J.. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov 2004;3(8):711–16. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

