Microbial Dark Matter Investigations: How Microbial Studies Transform Biological Knowledge and Empirically Sketch a Logic of Scientific Discovery
- PMID: 29420719
- PMCID: PMC5830969
- DOI: 10.1093/gbe/evy031
Microbial Dark Matter Investigations: How Microbial Studies Transform Biological Knowledge and Empirically Sketch a Logic of Scientific Discovery
Abstract
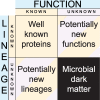
Microbes are the oldest and most widespread, phylogenetically and metabolically diverse life forms on Earth. However, they have been discovered only 334 years ago, and their diversity started to become seriously investigated even later. For these reasons, microbial studies that unveil novel microbial lineages and processes affecting or involving microbes deeply (and repeatedly) transform knowledge in biology. Considering the quantitative prevalence of taxonomically and functionally unassigned sequences in environmental genomics data sets, and that of uncultured microbes on the planet, we propose that unraveling the microbial dark matter should be identified as a central priority for biologists. Based on former empirical findings of microbial studies, we sketch a logic of discovery with the potential to further highlight the microbial unknowns.
Figures
References
-
- Barer MR, Harwood CR.. 1999. Bacterial viability and culturability. Adv Microb Physiol. 41:93–137. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

