Population Genetic Structure in Glyphosate-Resistant and -Susceptible Palmer Amaranth (Amaranthus palmeri) Populations Using Genotyping-by-sequencing (GBS)
- PMID: 29422910
- PMCID: PMC5788914
- DOI: 10.3389/fpls.2018.00029
Population Genetic Structure in Glyphosate-Resistant and -Susceptible Palmer Amaranth (Amaranthus palmeri) Populations Using Genotyping-by-sequencing (GBS)
Abstract
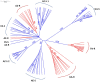
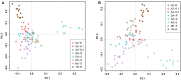
Palmer amaranth (Amaranthus palmeri) is a major weed in United States cotton and soybean production systems. Originally native to the Southwest, the species has spread throughout the country. In 2004 a population of A. palmeri was identified with resistance to glyphosate, a herbicide heavily relied on in modern no-tillage and transgenic glyphosate-resistant (GR) crop systems. This project aims to determine the degree of genetic relatedness among eight different populations of GR and glyphosate-susceptible (GS) A. palmeri from various geographic regions in the United States by analyzing patterns of phylogeography and diversity to ascertain whether resistance evolved independently or spread from outside to an Arizona locality (AZ-R). Shikimic acid accumulation and EPSPS genomic copy assays confirmed resistance or susceptibility. With a set of 1,351 single nucleotide polymorphisms (SNPs), discovered by genotyping-by-sequencing (GBS), UPGMA phylogenetic analysis, principal component analysis, Bayesian model-based clustering, and pairwise comparisons of genetic distances were conducted. A GR population from Tennessee and two GS populations from Georgia and Arizona were identified as genetically distinct while the remaining GS populations from Kansas, Arizona, and Nebraska clustered together with two GR populations from Arizona and Georgia. Within the latter group, AZ-R was most closely related to the GS populations from Kansas and Arizona followed by the GR population from Georgia. GR populations from Georgia and Tennessee were genetically distinct from each other. No isolation by distance was detected and A. palmeri was revealed to be a species with high genetic diversity. The data suggest the following two possible scenarios: either glyphosate resistance was introduced to the Arizona locality from the east, or resistance evolved independently in Arizona. Glyphosate resistance in the Georgia and Tennessee localities most likely evolved separately. Thus, modern farmers need to continue to diversify weed management practices and prevent seed dispersal to mitigate herbicide resistance evolution in A. palmeri.
Keywords: Palmer amaranth; SNP molecular markers; genetic relatedness; glyphosate; herbicide resistance; phylogeography; population genetics.
Figures




Similar articles
-
Glyphosate-resistant and glyphosate-susceptible Palmer amaranth (Amaranthus palmeri S. Wats.): hyperspectral reflectance properties of plants and potential for classification.Pest Manag Sci. 2014 Dec;70(12):1910-7. doi: 10.1002/ps.3755. Epub 2014 Mar 19. Pest Manag Sci. 2014. PMID: 24497403
-
First confirmation and characterization of target and non-target site resistance to glyphosate in Palmer amaranth (Amaranthus palmeri) from Mexico.Plant Physiol Biochem. 2017 Jun;115:212-218. doi: 10.1016/j.plaphy.2017.03.022. Epub 2017 Mar 31. Plant Physiol Biochem. 2017. PMID: 28384561
-
Survey of the genomic landscape surrounding the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene in glyphosate-resistant Amaranthus palmeri from geographically distant populations in the USA.Pest Manag Sci. 2018 May;74(5):1109-1117. doi: 10.1002/ps.4659. Epub 2017 Sep 11. Pest Manag Sci. 2018. PMID: 28686355
-
Deciphering the Mechanism of Glyphosate Resistance in Amaranthus palmeri by Cytogenomics.Cytogenet Genome Res. 2021;161(12):578-584. doi: 10.1159/000521409. Epub 2022 Jan 12. Cytogenet Genome Res. 2021. PMID: 35021177 Review.
-
History of Herbicide-Resistant Traits in Cotton in the U.S. and the Importance of Integrated Weed Management for Technology Stewardship.Plants (Basel). 2022 Apr 28;11(9):1189. doi: 10.3390/plants11091189. Plants (Basel). 2022. PMID: 35567190 Free PMC article. Review.
Cited by
-
High gene flow maintains genetic diversity following selection for high EPSPS copy number in the weed kochia (Amaranthaceae).Sci Rep. 2020 Nov 2;10(1):18864. doi: 10.1038/s41598-020-75345-6. Sci Rep. 2020. PMID: 33139774 Free PMC article.
-
Comparison of microbial community assemblages in the rhizosphere of three Amaranthus spp.PLoS One. 2023 Nov 29;18(11):e0294966. doi: 10.1371/journal.pone.0294966. eCollection 2023. PLoS One. 2023. PMID: 38019804 Free PMC article.
-
Genomic characterisation and dissection of the onset of resistance to acetyl CoA carboxylase-inhibiting herbicides in a large collection of Digitaria insularis from Brazil.Front Genet. 2024 Feb 19;15:1340852. doi: 10.3389/fgene.2024.1340852. eCollection 2024. Front Genet. 2024. PMID: 38440194 Free PMC article.
-
Population Genomic Approaches for Weed Science.Plants (Basel). 2019 Sep 19;8(9):354. doi: 10.3390/plants8090354. Plants (Basel). 2019. PMID: 31546893 Free PMC article. Review.
-
Homogeneity among glyphosate-resistant Amaranthus palmeri in geographically distant locations.PLoS One. 2020 Sep 9;15(9):e0233813. doi: 10.1371/journal.pone.0233813. eCollection 2020. PLoS One. 2020. PMID: 32903277 Free PMC article.
References
-
- Adamack A. T., Gruber B. (2014). PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol. Evol. 5 384–387. 10.1111/2041-210X.12158 - DOI
-
- Beard K. E., Lay K. S., Burton J. D., Burgos N., Lawton-Rauh A. L. (2014). Can Very Rapid Adaptation Arise without Ancestral Variation? Insight from the Molecular Evolution of Herbicide Resistance in Genus Amaranthus. Doctoral dissertation, Clemson University, Clemson, SC.
LinkOut - more resources
Full Text Sources
Other Literature Sources

