Mouse Obox and Crxos modulate preimplantation transcriptional profiles revealing similarity between paralogous mouse and human homeobox genes
- PMID: 29423137
- PMCID: PMC5787275
- DOI: 10.1186/s13227-018-0091-4
Mouse Obox and Crxos modulate preimplantation transcriptional profiles revealing similarity between paralogous mouse and human homeobox genes
Abstract
Background: ETCHbox genes are eutherian-specific homeobox genes expressed during preimplantation development at a time when the first cell lineage decisions are being made. The mouse has an unusual repertoire of ETCHbox genes with several gene families lost in evolution and the remaining two, Crxos and Obox, greatly divergent in sequence and number. Each has undergone duplication to give a double homeodomain Crxos locus and a large cluster of over 60 Obox loci. The gene content differences between species raise important questions about how evolution can tolerate loss of genes implicated in key developmental events.
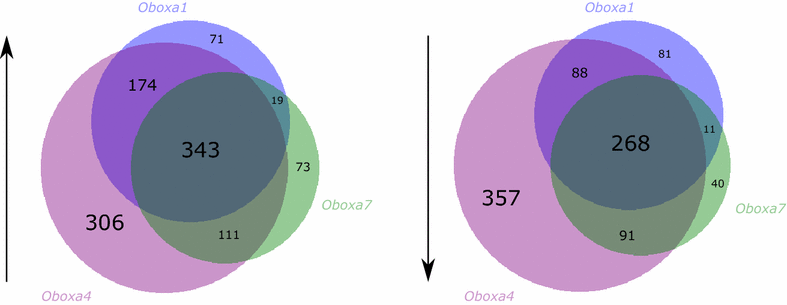
Results: We find that Crxos internal duplication occurred in the mouse lineage, while Obox duplication was stepwise, generating subgroups with distinct sequence and expression. Ectopic expression of three Obox genes and a Crxos transcript in primary mouse embryonic cells followed by transcriptome sequencing allowed investigation into their functional roles. We find distinct transcriptomic influences for different Obox subgroups and Crxos, including modulation of genes related to zygotic genome activation and preparation for blastocyst formation. Comparison with similar experiments performed using human homeobox genes reveals striking overlap between genes downstream of mouse Crxos and genes downstream of human ARGFX.
Conclusions: Mouse Crxos and human ARGFX homeobox genes are paralogous rather than orthologous, yet they have evolved to regulate a common set of genes. This suggests there was compensation of function alongside gene loss through co-option of a different locus. Functional compensation by non-orthologous genes with dissimilar sequences is unusual but may indicate underlying distributed robustness. Compensation may be driven by the strong evolutionary pressure for successful early embryo development.
Keywords: ARGFX; Blastocyst; Compensation; Gene duplication; Gene loss; Homeodomain; PRD class; Transcription factor.
Figures





References
-
- Holland PWH. Evolution of homeobox genes. Wiley Interdiscip Rev Dev Biol. 2013 - PubMed
-
- MacLean JA, II, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C, Wilkinson MF. Rhox: a new homeobox gene cluster. Cell Press. 2005 - PubMed
-
- Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL, Tracey A, Bobes RJ, Fragoso G, Sciutto E, Aslett M, Beasley H, Bennett HM, Cai J, Camicia F, Clark R, Cucher M, De Silva N, Day TA, Deplazes P, Estrada K, Fernandez C, Holland PWH. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013 - PMC - PubMed
-
- Booth HAF, Holland PWH. Annotation, nomenclature and evolution of four novel homeobox genes expressed in the human germ line. Gene. 2007 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

