Genetic diversity of equine herpesvirus 1 isolated from neurological, abortigenic and respiratory disease outbreaks
- PMID: 29423949
- PMCID: PMC5947664
- DOI: 10.1111/tbed.12809
Genetic diversity of equine herpesvirus 1 isolated from neurological, abortigenic and respiratory disease outbreaks
Abstract
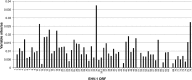
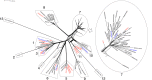
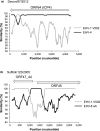
Equine herpesvirus 1 (EHV-1) causes respiratory disease, abortion, neonatal death and neurological disease in equines and is endemic in most countries. The viral factors that influence EHV-1 disease severity are poorly understood, and this has hampered vaccine development. However, the N752D substitution in the viral DNA polymerase catalytic subunit has been shown statistically to be associated with neurological disease. This has given rise to the term "neuropathic strain," even though strains lacking the polymorphism have been recovered from cases of neurological disease. To broaden understanding of EHV-1 diversity in the field, 78 EHV-1 strains isolated over a period of 35 years were sequenced. The great majority of isolates originated from the United Kingdom and included in the collection were low passage isolates from respiratory, abortigenic and neurological outbreaks. Phylogenetic analysis of regions spanning 80% of the genome showed that up to 13 viral clades have been circulating in the United Kingdom and that most of these are continuing to circulate. Abortion isolates grouped into nine clades, and neurological isolates grouped into five. Most neurological isolates had the N752D substitution, whereas most abortion isolates did not, although three of the neurological isolates from linked outbreaks had a different polymorphism. Finally, bioinformatic analysis suggested that recombination has occurred between EHV-1 clades, between EHV-1 and equine herpesvirus 4, and between EHV-1 and equine herpesvirus 8.
Keywords: equine herpesvirus type 1; sequencing, diversity.
© 2018 The Authors. Transboundary and Emerging Diseases Published by Blackwell Verlag GmbH.
Figures



References
-
- Allen, G. P. , & Breathnach, C. C. (2006). Quantification by real‐time PCR of the magnitude and duration of leucocyte‐associated viraemia in horses infected with neuropathogenic vs. non‐neuropathogenic strains of EHV‐1. Equine Veterinary Journal, 38, 252–257. - PubMed
-
- Allen, G. P. , Yeargan, M. R. , Turtinen, L. W. , Bryans, J. T. , & McCollum, W. H. (1983). Molecular epizootiologic studies of equine herpesvirus‐1 infections by restriction endonuclease fingerprinting of viral DNA. American Journal of Veterinary Research, 44, 263–271. - PubMed
-
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

