Genome analysis of clinical multilocus sequence Type 11 Klebsiella pneumoniae from China
- PMID: 29424684
- PMCID: PMC5857376
- DOI: 10.1099/mgen.0.000149
Genome analysis of clinical multilocus sequence Type 11 Klebsiella pneumoniae from China
Abstract
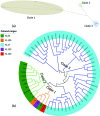
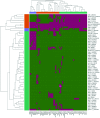
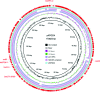
The increasing prevalence of KPC-producing Klebsiella pneumoniae strains in clinical settings has been largely attributed to dissemination of organisms of specific multilocus sequence types, such as ST258 and ST11. Compared with the ST258 clone, which is prevalent in North America and Europe, ST11 is common in China but information regarding its genetic features remains scarce. In this study, we performed detailed genetic characterization of ST11 K. pneumoniae strains by analyzing whole-genome sequences of 58 clinical strains collected from diverse geographic locations in China. The ST11 genomes were found to be highly heterogeneous and clustered into at least three major lineages based on the patterns of single-nucleotide polymorphisms. Exhibiting five different capsular types, these ST11 strains were found to harbor multiple resistance and virulence determinants such as the blaKPC-2 gene, which encodes carbapenemase, and the yersiniabactin-associated virulence genes irp, ybt and fyu. Moreover, genes encoding the virulence factor aerobactin and the regulator of the mucoid phenotype (rmpA) were detectable in six genomes, whereas genes encoding salmochelin were found in three genomes. In conclusion, our data indicated that carriage of a wide range of resistance and virulence genes constitutes the underlying basis of the high level of prevalence of ST11 in clinical settings. Such findings provide insight into the development of novel strategies for prevention, diagnosis and treatment of K. pneumoniae infections.
Keywords: Klebsiella pneumonia; ST11; ST258; antimicrobial resistance; cps loci; genome analysis; virulence.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

