Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus
- PMID: 29425101
- PMCID: PMC5892594
- DOI: 10.1074/jbc.RA118.001729
Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus
Abstract
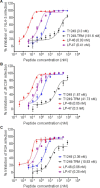
Enfuvirtide (T20) is the only viral fusion inhibitor approved for clinical use, but it has relatively weak anti-HIV activity and easily induces drug resistance. In succession to T20, T1249 has been designed as a 39-mer peptide composed of amino acid sequences derived from HIV-1, HIV-2, and simian immunodeficiency virus (SIV); however, its development has been suspended due to formulation difficulties. We recently developed a T20-based lipopeptide (LP-40) showing greatly improved pharmaceutical properties. Here, we generated a T1249-based lipopeptide, termed LP-46, by replacing its C-terminal tryptophan-rich sequence with fatty acid. As compared with T20, T1249, and LP-40, the truncated LP-46 (31-mer) had dramatically increased activities in inhibiting a large panel of HIV-1 subtypes, with IC50 values approaching low picomolar concentrations. Also, LP-46 was an exceptionally potent inhibitor against HIV-2, SIV, and T20-resistant variants, and it displayed obvious synergistic effects with LP-40. Furthermore, we showed that LP-46 had increased helical stability and binding affinity with the target site. The crystal structure of LP-46 in complex with a target surrogate revealed its critical binding motifs underlying the mechanism of action. Interestingly, it was found that the introduced pocket-binding domain in LP-46 did not interact with the gp41 pocket as expected; instead, it adopted a mode similar to that of LP-40. Therefore, our studies have provided an exceptionally potent and broad fusion inhibitor for developing new anti-HIV drugs, which can also serve as a tool to exploit the mechanisms of viral fusion and inhibition.
Keywords: HIV-1; HIV-2; T1249; T20; antiviral agent; crystal structure; fusion inhibitor; human immunodeficiency virus (HIV); inhibitor; lipopeptide; membrane fusion; peptides; structure-function.
© 2018 Zhu et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



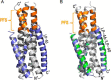

Similar articles
-
Enfuvirtide (T20)-Based Lipopeptide Is a Potent HIV-1 Cell Fusion Inhibitor: Implications for Viral Entry and Inhibition.J Virol. 2017 Aug 24;91(18):e00831-17. doi: 10.1128/JVI.00831-17. Print 2017 Sep 15. J Virol. 2017. PMID: 28659478 Free PMC article.
-
Design and Characterization of Cholesterylated Peptide HIV-1/2 Fusion Inhibitors with Extremely Potent and Long-Lasting Antiviral Activity.J Virol. 2019 May 15;93(11):e02312-18. doi: 10.1128/JVI.02312-18. Print 2019 Jun 1. J Virol. 2019. PMID: 30867304 Free PMC article.
-
A Lipopeptide HIV-1/2 Fusion Inhibitor with Highly Potent In Vitro, Ex Vivo, and In Vivo Antiviral Activity.J Virol. 2017 May 12;91(11):e00288-17. doi: 10.1128/JVI.00288-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28356533 Free PMC article.
-
Synthesized peptide inhibitors of HIV-1 gp41-dependent membrane fusion.Curr Pharm Des. 2013;19(10):1800-9. doi: 10.2174/1381612811319100004. Curr Pharm Des. 2013. PMID: 23092277 Review.
-
Inhibition of HIV-1 by fusion inhibitors.Curr Pharm Des. 2010;16(33):3716-28. doi: 10.2174/138161210794079218. Curr Pharm Des. 2010. PMID: 21128887 Review.
Cited by
-
Recent advances in developing small-molecule inhibitors against SARS-CoV-2.Acta Pharm Sin B. 2022 Apr;12(4):1591-1623. doi: 10.1016/j.apsb.2021.06.016. Epub 2021 Jul 2. Acta Pharm Sin B. 2022. PMID: 34249607 Free PMC article. Review.
-
The conformational epitope of a gp41-specific mucosal protective IgA binds to the HIV-1 envelope and neutralizes infection.Acta Pharmacol Sin. 2025 Jun 5. doi: 10.1038/s41401-025-01535-5. Online ahead of print. Acta Pharmacol Sin. 2025. PMID: 40473821
-
A Novel CXCR4 Targeting Protein SDF-1/54 as an HIV-1 Entry Inhibitor.Viruses. 2019 Sep 18;11(9):874. doi: 10.3390/v11090874. Viruses. 2019. PMID: 31540474 Free PMC article.
-
Current and Future Therapeutic Strategies for Lentiviral Eradication from Macrophage Reservoirs.J Neuroimmune Pharmacol. 2019 Mar;14(1):68-93. doi: 10.1007/s11481-018-9814-5. Epub 2018 Oct 13. J Neuroimmune Pharmacol. 2019. PMID: 30317409 Free PMC article. Review.
-
Design of Potent Membrane Fusion Inhibitors against SARS-CoV-2, an Emerging Coronavirus with High Fusogenic Activity.J Virol. 2020 Jul 1;94(14):e00635-20. doi: 10.1128/JVI.00635-20. Print 2020 Jul 1. J Virol. 2020. PMID: 32376627 Free PMC article.
References
-
- Yahi N., Fantini J., Baghdiguian S., Mabrouk K., Tamalet C., Rochat H., Van Rietschoten J., and Sabatier J. M. (1995) SPC3, a synthetic peptide derived from the V3 domain of human immunodeficiency virus type 1 (HIV-1) gp120, inhibits HIV-1 entry into CD4+ and CD4− cells by two distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 92, 4867–4871 10.1073/pnas.92.11.4867 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

