Resistance analysis of genotype 3 hepatitis C virus indicates subtypes inherently resistant to nonstructural protein 5A inhibitors
- PMID: 29425396
- PMCID: PMC6492296
- DOI: 10.1002/hep.29837
Resistance analysis of genotype 3 hepatitis C virus indicates subtypes inherently resistant to nonstructural protein 5A inhibitors
Abstract
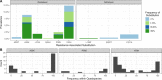
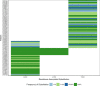
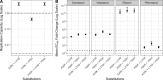
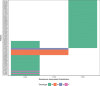
Hepatitis C virus (HCV) genotype (gt) 3 is highly prevalent globally, with non-gt3a subtypes common in Southeast Asia. Resistance-associated substitutions (RASs) have been shown to play a role in treatment failure. However, the role of RASs in gt3 is not well understood. We report the prevalence of RASs in a cohort of direct-acting antiviral treatment-naive, gt3-infected patients, including those with rarer subtypes, and evaluate the effect of these RASs on direct-acting antivirals in vitro. Baseline samples from 496 gt3 patients enrolled in the BOSON clinical trial were analyzed by next-generation sequencing after probe-based enrichment for HCV. Whole viral genomes were analyzed for the presence of RASs to approved direct-acting antivirals. The resistance phenotype of RASs in combination with daclatasvir, velpatasvir, pibrentasvir, elbasvir, and sofosbuvir was measured using the S52 ΔN gt3a replicon model. The nonstructural protein 5A A30K and Y93H substitutions were the most common at 8.9% (n = 44) and 12.3% (n = 61), respectively, and showed a 10-fold and 11-fold increase in 50% effect concentration for daclatasvir compared to the unmodified replicon. Paired RASs (A30K + L31M and A30K + Y93H) were identified in 18 patients (9 of each pair); these combinations were shown to be highly resistant to daclatasvir, velpatasvir, elbasvir, and pibrentasvir. The A30K + L31M combination was found in all gt3b and gt3g samples. Conclusion: Our study reveals high frequencies of RASs to nonstructural protein 5A inhibitors in gt3 HCV; the paired A30K + L31M substitutions occur in all patients with gt3b and gt3g virus, and in vitro analysis suggests that these subtypes may be inherently resistant to all approved nonstructural protein 5A inhibitors for gt3 HCV. (Hepatology 2018).
© 2018 The Authors. Hepatology published by Wiley Periodicals, Inc. on behalf of American Association for the Study of Liver Diseases.
Figures



References
-
- World Health Organization . Global hepatitis report, 2017. Geneva, Switzerland: World Health Organization; 2017.
-
- Kowdley KV, Gordon SC, Reddy KR, Rossaro L, Bernstein DE, Lawitz E, et al. Ledipasvir and sofosbuvir for 8 or 12 weeks for chronic HCV without cirrhosis. N Engl J Med 2014;370:1879‐1888. - PubMed
-
- Feld JJ, Jacobson IM, Hézode C, Asselah T, Ruane PJ, Gruener N, et al. Sofosbuvir and velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N Engl J Med 2015;373:2599‐2607. - PubMed
-
- Sulkowski MS, Gardiner DF, Rodriguez‐Torres M, Reddy KR, Hassanein T, Jacobson I, et al. Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N Engl J Med 2014;370:211‐221. - PubMed
-
- Wyles DL, Ruane PJ, Sulkowski MS, Dieterich D, Luetkemeyer A, Morgan TR, et al. Daclatasvir plus sofosbuvir for HCV in patients coinfected with HIV‐1. N Engl J Med 2015;373:714‐725. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

