Dysbiosis of Inferior Turbinate Microbiota Is Associated with High Total IgE Levels in Patients with Allergic Rhinitis
- PMID: 29426044
- PMCID: PMC5865026
- DOI: 10.1128/IAI.00934-17
Dysbiosis of Inferior Turbinate Microbiota Is Associated with High Total IgE Levels in Patients with Allergic Rhinitis
Abstract
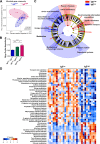
Abnormalities in the human microbiota are associated with the etiology of allergic diseases. Although disease site-specific microbiota may be associated with disease pathophysiology, the role of the nasal microbiota is unclear. We sought to characterize the microbiota of the site of allergic rhinitis, the inferior turbinate, in subjects with allergic rhinitis (n = 20) and healthy controls (n = 12) and to examine the relationship of mucosal microbiota with disease occurrence, sensitized allergen number, and allergen-specific and total IgE levels. Microbial dysbiosis correlated significantly with total IgE levels representing combined allergic responses but not with disease occurrence, the number of sensitized allergens, or house dust mite allergen-specific IgE levels. Compared to the populations in individuals with low total IgE levels (group IgElow), low microbial biodiversity with a high relative abundance of Firmicutes phylum (Staphylococcus aureus) and a low relative abundance of Actinobacteria phylum (Propionibacterium acnes) was observed in individuals with high total serum IgE levels (group IgEhigh). Phylogeny-based microbial functional potential predicted by the 16S rRNA gene indicated an increase in signal transduction-related genes and a decrease in energy metabolism-related genes in group IgEhigh as shown in the microbial features with atopic and/or inflammatory diseases. Thus, dysbiosis of the inferior turbinate mucosa microbiota, particularly an increase in S. aureus and a decrease in P. acnes, is linked to high total IgE levels in allergic rhinitis, suggesting that inferior turbinate microbiota may be affected by accumulated allergic responses against sensitized allergens and that site-specific microbial alterations play a potential role in disease pathophysiology.
Keywords: Propionibacterium acnes; Staphylococcus aureus; allergic rhinitis; commensal microbiota; immunoglobulin E; inferior turbinate; multiple-allergen simultaneous test; nasal mucosa; skin prick test.
Copyright © 2018 American Society for Microbiology.
Figures



References
-
- Cruz AA, Bousquet J, Khaltaev N. 2007. Global surveillance, prevention and control of chronic respiratory diseases: a comprehensive approach. World Health Organization, Geneva, Switzerland.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

