Identifying source populations for the reintroduction of the Eurasian beaver, Castor fiber L. 1758, into Britain: evidence from ancient DNA
- PMID: 29426903
- PMCID: PMC5807398
- DOI: 10.1038/s41598-018-21173-8
Identifying source populations for the reintroduction of the Eurasian beaver, Castor fiber L. 1758, into Britain: evidence from ancient DNA
Abstract
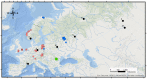
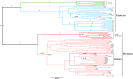
Establishing true phylogenetic relationships between populations is a critical consideration when sourcing individuals for translocation. This presents huge difficulties with threatened and endangered species that have become extirpated from large areas of their former range. We utilise ancient DNA (aDNA) to reconstruct the phylogenetic relationships of a keystone species which has become extinct in Britain, the Eurasian beaver Castor fiber. We sequenced seventeen 492 bp partial tRNAPro and control region sequences from Late Pleistocene and Holocene age beavers and included these in network, demographic and genealogy analyses. The mode of postglacial population expansion from refugia was investigated by employing tests of neutrality and a pairwise mismatch distribution analysis. We found evidence of a pre-Late Glacial Maximum ancestor for the Western C. fiber clade which experienced a rapid demographic expansion during the terminal Pleistocene to early Holocene period. Ancient British beavers were found to originate from the Western phylogroup but showed no phylogenetic affinity to any one modern relict population over another. Instead, we find that they formed part of a large, continuous, pan-Western European clade that harbored little internal substructure. Our study highlights the utility of aDNA in reconstructing population histories of extirpated species which has real-world implications for conservation planning.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Ancient mitochondrial DNA and the genetic history of Eurasian beaver (Castor fiber) in Europe.Mol Ecol. 2014 Apr;23(7):1717-29. doi: 10.1111/mec.12691. Mol Ecol. 2014. PMID: 24795996
-
Mitochondrial genomes reveal slow rates of molecular evolution and the timing of speciation in beavers (Castor), one of the largest rodent species.PLoS One. 2011 Jan 28;6(1):e14622. doi: 10.1371/journal.pone.0014622. PLoS One. 2011. PMID: 21307956 Free PMC article.
-
The genetic legacy of multiple beaver reintroductions in Central Europe.PLoS One. 2014 May 14;9(5):e97619. doi: 10.1371/journal.pone.0097619. eCollection 2014. PLoS One. 2014. PMID: 24827835 Free PMC article.
-
Time to Spread Your Wings: A Review of the Avian Ancient DNA Field.Genes (Basel). 2017 Jul 18;8(7):184. doi: 10.3390/genes8070184. Genes (Basel). 2017. PMID: 28718817 Free PMC article. Review.
-
Beaver: Nature's ecosystem engineers.WIREs Water. 2021 Jan-Feb;8(1):e1494. doi: 10.1002/wat2.1494. Epub 2020 Nov 27. WIREs Water. 2021. PMID: 33614026 Free PMC article. Review.
Cited by
-
Archaeological mitogenomes illuminate the historical ecology of sea otters (Enhydra lutris) and the viability of reintroduction.Proc Biol Sci. 2020 Dec 9;287(1940):20202343. doi: 10.1098/rspb.2020.2343. Epub 2020 Dec 2. Proc Biol Sci. 2020. PMID: 33259759 Free PMC article.
-
Population genetic structure of Texas horned lizards: implications for reintroduction and captive breeding.PeerJ. 2019 Oct 1;7:e7746. doi: 10.7717/peerj.7746. eCollection 2019. PeerJ. 2019. PMID: 31592183 Free PMC article.
-
Genetic Diversity and Phylogenetic Relationships of Castor fiber birulai in Xinjiang, China, Revealed by Mitochondrial Cytb and D-loop Sequence Analyses.Animals (Basel). 2025 Jul 16;15(14):2096. doi: 10.3390/ani15142096. Animals (Basel). 2025. PMID: 40723559 Free PMC article.
References
-
- Houde ALS, Garner SR, Neff BD. Restoring species through reintroductions: strategies for source population selection. Restor. Ecol. 2015;23:746–753. doi: 10.1111/rec.12280. - DOI
-
- Fischer J, Lindenmayer DB. An assessment of the published results of animal relocations. Biol. Conserv. 2000;96:1–11. doi: 10.1016/S0006-3207(00)00048-3. - DOI
-
- IUCN/SSC. Guidelines for reintroductions and other conservation translocations. Version 1.0. IUCN Species Survival Commission, Gland, Switzerland, viiii+57pp. https://portals.iucn.org/library/efiles/documents/2013-009.pdf (2013).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

