TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing
- PMID: 29426904
- PMCID: PMC5807518
- DOI: 10.1038/s41467-018-02936-3
TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing
Abstract
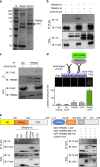
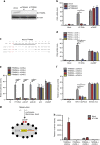
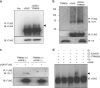
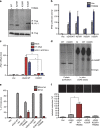
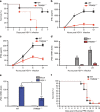
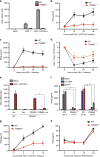
Intracellular nucleic acid sensors often undergo sophisticated modifications that are critical for the regulation of antimicrobial responses. Upon recognition of DNA, the cytosolic sensor cyclic GMP-AMP (cGAMP) synthase (cGAS) produces the second messenger cGAMP, which subsequently initiates downstream signaling to induce interferon-αβ (IFNαβ) production. Here we report that TRIM56 E3 ligase-induced monoubiquitination of cGAS is important for cytosolic DNA sensing and IFNαβ production to induce anti-DNA viral immunity. TRIM56 induces the Lys335 monoubiquitination of cGAS, resulting in a marked increase of its dimerization, DNA-binding activity, and cGAMP production. Consequently, TRIM56-deficient cells are defective in cGAS-mediated IFNαβ production upon herpes simplex virus-1 (HSV-1) infection. Furthermore, TRIM56-deficient mice show impaired IFNαβ production and high susceptibility to lethal HSV-1 infection but not to influenza A virus infection. This adds TRIM56 as a crucial component of the cytosolic DNA sensing pathway that induces anti-DNA viral innate immunity.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

