Tridimensional visualization reveals direct communication between the embryo and glands critical for implantation
- PMID: 29426931
- PMCID: PMC5807548
- DOI: 10.1038/s41467-018-03092-4
Tridimensional visualization reveals direct communication between the embryo and glands critical for implantation
Abstract
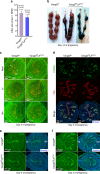
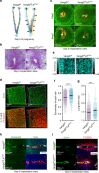
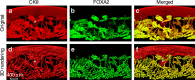
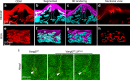
Embryo implantation is central to pregnancy success. Our previous understanding is limited by studying this phenomenon primarily in two dimensions. Here we employ 3D visualization, revealing that epithelial evaginations that form implantation chambers (crypts) consistently arise with preexisting glands, suggesting direct access of glands to embryos within the chamber. While the lobular domains of the glands become more developed, the ductal regions continue to elongate and progressively stretch following implantation. Using diapausing mice and mice with deletion of the planar cell polarity gene Vangl2 in uterine epithelial cells, we show that dynamic changes in gland topography depend on implantation-competent blastocysts and planar cell polarity. By transferring blastocyst-size beads preloaded with HB-EGF in pseudopregnant mice, we found that HB-EGF is a trigger for the communication between embryos and glands. Glands directly connecting the crypt encasing the embryo during implantation are therefore fundamental to pregnancy success.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Song H, et al. Cytosolic phospholipase A2alpha is crucial [correction of A2alpha deficiency is crucial] for ‘on-time’ embryo implantation that directs subsequent development. Development. 2002;129:2879–2889. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

