Genetic alterations driving metastatic colony formation are acquired outside of the primary tumour in melanoma
- PMID: 29426936
- PMCID: PMC5807512
- DOI: 10.1038/s41467-017-02674-y
Genetic alterations driving metastatic colony formation are acquired outside of the primary tumour in melanoma
Abstract
Mouse models indicate that metastatic dissemination occurs extremely early; however, the timing in human cancers is unknown. We therefore determined the time point of metastatic seeding relative to tumour thickness and genomic alterations in melanoma. Here, we find that lymphatic dissemination occurs shortly after dermal invasion of the primary lesion at a median thickness of ~0.5 mm and that typical driver changes, including BRAF mutation and gained or lost regions comprising genes like MET or CDKNA2, are acquired within the lymph node at the time of colony formation. These changes define a colonisation signature that was linked to xenograft formation in immunodeficient mice and death from melanoma. Thus, melanoma cells leave primary tumours early and evolve at different sites in parallel. We propose a model of metastatic melanoma dormancy, evolution and colonisation that will inform direct monitoring of adjuvant therapy targets.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
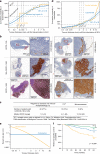



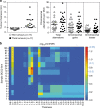

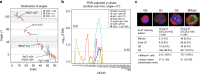

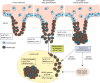
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

