Transformation of Summary Statistics from Linear Mixed Model Association on All-or-None Traits to Odds Ratio
- PMID: 29429966
- PMCID: PMC5887138
- DOI: 10.1534/genetics.117.300360
Transformation of Summary Statistics from Linear Mixed Model Association on All-or-None Traits to Odds Ratio
Abstract
Genome-wide association studies (GWAS) have identified thousands of loci that are robustly associated with complex diseases. The use of linear mixed model (LMM) methodology for GWAS is becoming more prevalent due to its ability to control for population structure and cryptic relatedness and to increase power. The odds ratio (OR) is a common measure of the association of a disease with an exposure (e.g., a genetic variant) and is readably available from logistic regression. However, when the LMM is applied to all-or-none traits it provides estimates of genetic effects on the observed 0-1 scale, a different scale to that in logistic regression. This limits the comparability of results across studies, for example in a meta-analysis, and makes the interpretation of the magnitude of an effect from an LMM GWAS difficult. In this study, we derived transformations from the genetic effects estimated under the LMM to the OR that only rely on summary statistics. To test the proposed transformations, we used real genotypes from two large, publicly available data sets to simulate all-or-none phenotypes for a set of scenarios that differ in underlying model, disease prevalence, and heritability. Furthermore, we applied these transformations to GWAS summary statistics for type 2 diabetes generated from 108,042 individuals in the UK Biobank. In both simulation and real-data application, we observed very high concordance between the transformed OR from the LMM and either the simulated truth or estimates from logistic regression. The transformations derived and validated in this study improve the comparability of results from prospective and already performed LMM GWAS on complex diseases by providing a reliable transformation to a common comparative scale for the genetic effects.
Keywords: OR; complex diseases; genome-wide association studies; linear mixed models; summary statistics.
Copyright © 2018 by the Genetics Society of America.
Figures
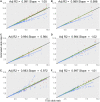
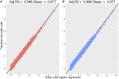
References
-
- Aldrich J. H., Nelson F. D., 1984. Linear Probability, Logit, and Probit Models, Vol. 45 Sage, London.
-
- Chang B.-H., Lipsitz S., Waternaux C., 2000. Logistic regression in meta-analysis using aggregate data. J. Appl. Stat. 27: 411–424.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

