Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity
- PMID: 29432122
- PMCID: PMC5839769
- DOI: 10.1084/jem.20172222
Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity
Abstract
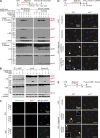
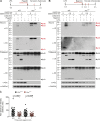
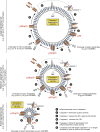
Host-protective caspase-1 activity must be tightly regulated to prevent pathology, but mechanisms controlling the duration of cellular caspase-1 activity are unknown. Caspase-1 is activated on inflammasomes, signaling platforms that facilitate caspase-1 dimerization and autoprocessing. Previous studies with recombinant protein identified a caspase-1 tetramer composed of two p20 and two p10 subunits (p20/p10) as an active species. In this study, we report that in the cell, the dominant species of active caspase-1 dimers elicited by inflammasomes are in fact full-length p46 and a transient species, p33/p10. Further p33/p10 autoprocessing occurs with kinetics specified by inflammasome size and cell type, and this releases p20/p10 from the inflammasome, whereupon the tetramer becomes unstable in cells and protease activity is terminated. The inflammasome-caspase-1 complex thus functions as a holoenzyme that directs the location of caspase-1 activity but also incorporates an intrinsic self-limiting mechanism that ensures timely caspase-1 deactivation. This intrinsic mechanism of inflammasome signal shutdown offers a molecular basis for the transient nature, and coordinated timing, of inflammasome-dependent inflammatory responses.
© 2018 Boucher et al.
Figures




Comment in
-
Defusing inflammasomes.J Exp Med. 2018 Mar 5;215(3):723-724. doi: 10.1084/jem.20180241. Epub 2018 Feb 12. J Exp Med. 2018. PMID: 29440361 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

