Lipid bilayer composition modulates the unfolding free energy of a knotted α-helical membrane protein
- PMID: 29432185
- PMCID: PMC5828594
- DOI: 10.1073/pnas.1714668115
Lipid bilayer composition modulates the unfolding free energy of a knotted α-helical membrane protein
Abstract
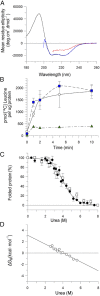
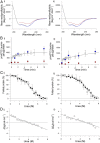
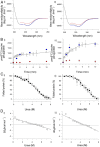
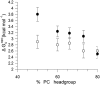
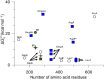
α-Helical membrane proteins have eluded investigation of their thermodynamic stability in lipid bilayers. Reversible denaturation curves have enabled some headway in determining unfolding free energies. However, these parameters have been limited to detergent micelles or lipid bicelles, which do not possess the same mechanical properties as lipid bilayers that comprise the basis of natural membranes. We establish reversible unfolding of the membrane transporter LeuT in lipid bilayers, enabling the comparison of apparent unfolding free energies in different lipid compositions. LeuT is a bacterial ortholog of neurotransmitter transporters and contains a knot within its 12-transmembrane helical structure. Urea is used as a denaturant for LeuT in proteoliposomes, resulting in the loss of up to 30% helical structure depending upon the lipid bilayer composition. Urea unfolding of LeuT in liposomes is reversible, with refolding in the bilayer recovering the original helical structure and transport activity. A linear dependence of the unfolding free energy on urea concentration enables the free energy to be extrapolated to zero denaturant. Increasing lipid headgroup charge or chain lateral pressure increases the thermodynamic stability of LeuT. The mechanical and charge properties of the bilayer also affect the ability of urea to denature the protein. Thus, we not only gain insight to the long-sought-after thermodynamic stability of an α-helical protein in a lipid bilayer but also provide a basis for studies of the folding of knotted proteins in a membrane environment.
Keywords: membrane proteins; protein folding; protein–lipid interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
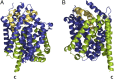
References
-
- Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. - PubMed
-
- Booth PJ. Sane in the membrane: Designing systems to modulate membrane proteins. Curr Opin Struct Biol. 2005;15:435–440. - PubMed
-
- Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem. 2000;69:881–922. - PubMed
-
- Engelman DM, et al. Membrane protein folding: Beyond the two stage model. FEBS Lett. 2003;555:122–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

