Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection
- PMID: 29433440
- PMCID: PMC5809863
- DOI: 10.1186/s12866-017-1148-6
Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection
Abstract
Background: Hypervirulent K. pneumoniae (hvKp) causes severe community acquired infections, predominantly in Asia. Though initially isolated from liver abscesses, they are now prevalent among invasive infections such as bacteraemia. There have been no studies reported till date on the prevalence and characterisation of hvKp in India. The objective of this study is to characterise the hypervirulent strains isolated from bacteraemic patients for determination of various virulence genes and resistance genes and also to investigate the difference between healthcare associated and community acquired hvKp with respect to clinical profile, antibiogram, clinical outcome and molecular epidemiology.
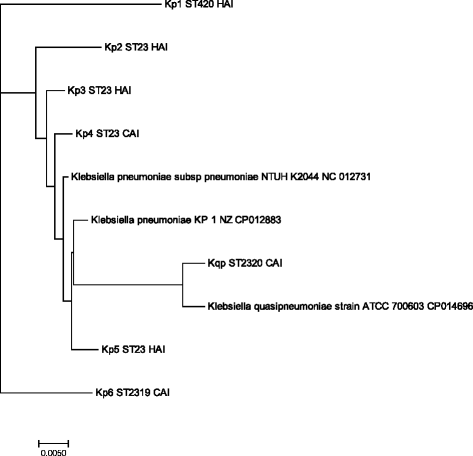
Results: Seven isolates that were susceptible to all of the first and second line antimicrobials and phenotypically identified by positive string test were included in the study. They were then confirmed genotypically by presence of rmpA and rmpA2 by PCR. Among the study isolates, four were from patients with healthcare associated infections; none were fatal. All patients with community acquired infection possessed chronic liver disease with fatal outcome. Genes encoding for siderophores such as aerobactin, enterobactin, yersiniabactin, allantoin metabolism and iron uptake were identified by whole genome sequencing. Five isolates belonged to K1 capsular type including one K. quasipneumoniae. None belonged to K2 capsular type. Four isolates belonged to the international clone ST23 among which three were health-care associated and possessed increased virulence genes. Two novel sequence types were identified in the study; K. pneumoniae belonging to ST2319 and K. quasipneumoniae belonging to ST2320. Seventh isolate belonged to ST420.
Conclusion: This is the first report on whole genome analysis of hypervirulent K. pneumoniae from India. The novel sequence types described in this study indicate that these strains are evolving and hvKp is now spread across various clonal types. Studies to monitor the prevalence of hvKp is needed since there is a potential for the community acquired isolates to develop multidrug resistance in hospital environment and may pose a major challenge for clinical management.
Keywords: Bacteremia; Hypervirulent; K. pneumoniae; K. quasipneumoniae; Virulence factors; Whole genome sequencing.
Conflict of interest statement
Ethics approval and consent to participate
This study was approved by the ethical committee of Christian Medical College, Vellore, India. This is a retrospective study in which the isolates are used without the patient identifier and hence patient consent was not obtained.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Brisse S, Passet V, Grimont PA. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol. 2014;64(9):3146–3152. doi: 10.1099/ijs.0.062737-0. - DOI - PubMed
-
- Zhang Y, Zhao C, Wang Q, Wang X, Chen H, Li H, Zhang F, Li S, Wang R, Wang H. High prevalence of Hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60(10):6115–6120. doi: 10.1128/AAC.01127-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

