SMURF1 facilitates estrogen receptor ɑ signaling in breast cancer cells
- PMID: 29433542
- PMCID: PMC5808446
- DOI: 10.1186/s13046-018-0672-z
SMURF1 facilitates estrogen receptor ɑ signaling in breast cancer cells
Abstract
Background: Estrogen receptor alpha (ER alpha) is expressed in the majority of breast cancers and promotes estrogen-dependent cancer progression. ER alpha positive breast cancer can be well controlled by ER alpha modulators, such as tamoxifen. However, tamoxifen resistance is commonly observed by altered ER alpha signaling. Thus, further understanding of the molecular mechanisms, which regulates ER alpha signaling, is important to improve breast cancer therapy.
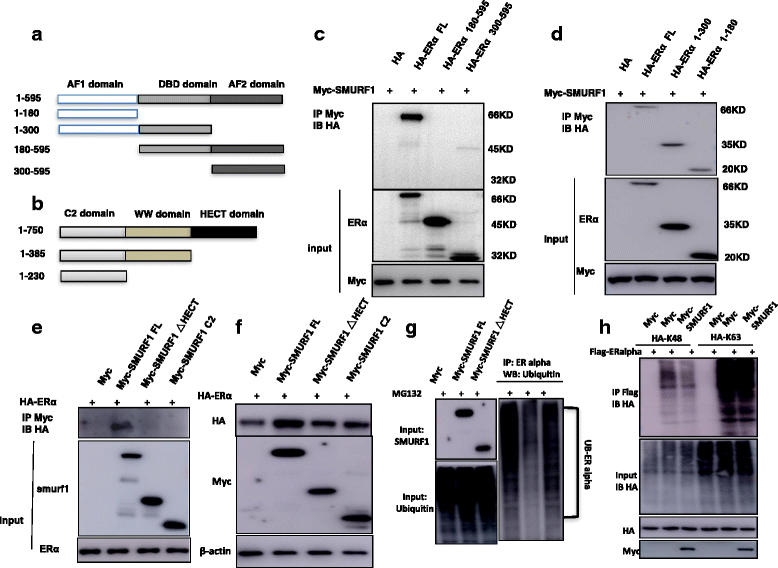
Methods: SMURF1 and ER alpha protein expression levels were measured by western blot, while the ER alpha target genes were measured by real-time PCR. WST-1 assay was used to measure cell viability; the xeno-graft tumor model were used for in vivo study. RNA sequencing was analyzed by Ingenuity Pathway Analysis. Identification of ER alpha signaling was accomplished with luciferase assays, real-time RT-PCR and Western blotting. Protein stability assay and ubiquitin assay was used to detect ER alpha protein degradation. Immuno-precipitation based assays were used to detect the interaction domain between ER alpha and SMURF1. The ubiquitin-based Immuno-precipitation based assays were used to detect the specific ubiquitination manner happened on ER alpha.
Results: Here, we identify the E3 ligase SMURF1 facilitates ER alpha signaling. We show that depletion SMURF1 decreases ER alpha positive cell proliferation in vitro and in vivo. SMURF1 depletion based RNA-sequence data shows SMURF1 is necessary for ER alpha target gene expression in the transcriptomic scale. Immunoprecipitation indicates that SMURF1 associates with the N-terminal of ER alpha in the cytoplasm via its HECT domain. SMURF1 increases ER alpha stability, possibly by inhibiting K48-specific poly-ubiquitination process on ER alpha protein. Interestingly, SMURF1 expression could be induced via estradiol treatment.
Conclusions: Our study reveals a novel positive feedback between SMURF1 and ER alpha signaling in supporting breast cancer growth. Targeting SMURF1 could be one promising strategy for ER alpha positive breast cancer treatment.
Keywords: Breast cancer; ER alpha; Protein stability; SMURF1; Ubiquitination.
Conflict of interest statement
Ethics approval and consent to participate
This study was reviewed and approved by the Ethical Board at Xinxiang Medical University.
Consent for publication
Not applicable.
Competing interests
The authors’ declare that they have no competing of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
-
- Breast International Group 1-98 Collaborative G. Thurlimann B, Keshaviah A, Coates AS, Mouridsen H, Mauriac L, Forbes JF, Paridaens R, Castiglione-Gertsch M, Gelber RD, et al. A comparison of letrozole and tamoxifen in postmenopausal women with early breast cancer. N Engl J Med. 2005;353(26):2747–2757. doi: 10.1056/NEJMoa052258. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- No. 8170110153/National Science Foundation for Young Scientists of China
- No.U1604190/the Joint Fund of the National Natural Science Foundation of China
- No.15IRTSTHN025/the Program for Innovative Research Team (in Science and Technology) in University of Henan Province
- 2012AA02A201-1/the National High Technology Research and Development Program of China
- No.17A310025/the Foundation of Henan Educational Committee
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

