Poldip2 is an oxygen-sensitive protein that controls PDH and αKGDH lipoylation and activation to support metabolic adaptation in hypoxia and cancer
- PMID: 29434038
- PMCID: PMC5828627
- DOI: 10.1073/pnas.1720693115
Poldip2 is an oxygen-sensitive protein that controls PDH and αKGDH lipoylation and activation to support metabolic adaptation in hypoxia and cancer
Abstract
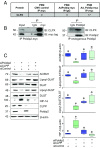
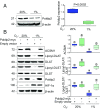
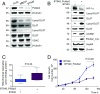
Although the addition of the prosthetic group lipoate is essential to the activity of critical mitochondrial catabolic enzymes, its regulation is unknown. Here, we show that lipoylation of the pyruvate dehydrogenase and α-ketoglutarate dehydrogenase (αKDH) complexes is a dynamically regulated process that is inhibited under hypoxia and in cancer cells to restrain mitochondrial respiration. Mechanistically, we found that the polymerase-δ interacting protein 2 (Poldip2), a nuclear-encoded mitochondrial protein of unknown function, controls the lipoylation of the pyruvate and α-KDH dihydrolipoamide acetyltransferase subunits by a mechanism that involves regulation of the caseinolytic peptidase (Clp)-protease complex and degradation of the lipoate-activating enzyme Ac-CoA synthetase medium-chain family member 1 (ACSM1). ACSM1 is required for the utilization of lipoic acid derived from a salvage pathway, an unacknowledged lipoylation mechanism. In Poldip2-deficient cells, reduced lipoylation represses mitochondrial function and induces the stabilization of hypoxia-inducible factor 1α (HIF-1α) by loss of substrate inhibition of prolyl-4-hydroxylases (PHDs). HIF-1α-mediated retrograde signaling results in a metabolic reprogramming that resembles hypoxic and cancer cell adaptation. Indeed, we observe that Poldip2 expression is down-regulated by hypoxia in a variety of cell types and basally repressed in triple-negative cancer cells, leading to inhibition of lipoylation of the pyruvate and α-KDH complexes and mitochondrial dysfunction. Increasing mitochondrial lipoylation by forced expression of Poldip2 increases respiration and reduces the growth rate of cancer cells. Our work unveils a regulatory mechanism of catabolic enzymes required for metabolic plasticity and highlights the role of Poldip2 as key during hypoxia and cancer cell metabolic adaptation.
Keywords: Poldip2; hypoxia; lipoylation; metabolism; mitochondria.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Comment in
-
Reply to Bailey et al.: New perspectives on the novel role of the Poldip2/ACSM1 axis in a functional mammalian lipoylation salvage pathway.Proc Natl Acad Sci U S A. 2018 Aug 7;115(32):E7460-E7461. doi: 10.1073/pnas.1807968115. Epub 2018 Jul 24. Proc Natl Acad Sci U S A. 2018. PMID: 30042216 Free PMC article. No abstract available.
-
Different opinion on the reported role of Poldip2 and ACSM1 in a mammalian lipoic acid salvage pathway controlling HIF-1 activation.Proc Natl Acad Sci U S A. 2018 Aug 7;115(32):E7458-E7459. doi: 10.1073/pnas.1804041115. Epub 2018 Jul 24. Proc Natl Acad Sci U S A. 2018. PMID: 30042217 Free PMC article. No abstract available.
References
-
- Liu L, Rodriguez-Belmonte EM, Mazloum N, Xie B, Lee MY. Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen. J Biol Chem. 2003;278:10041–10047. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

