Absence of warmth permits epigenetic memory of winter in Arabidopsis
- PMID: 29434233
- PMCID: PMC5809604
- DOI: 10.1038/s41467-018-03065-7
Absence of warmth permits epigenetic memory of winter in Arabidopsis
Abstract
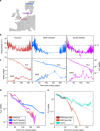
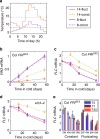
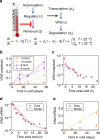
Plants integrate widely fluctuating temperatures to monitor seasonal progression. Here, we investigate the temperature signals in field conditions that result in vernalisation, the mechanism by which flowering is aligned with spring. We find that multiple, distinct aspects of the temperature profile contribute to vernalisation. In autumn, transient cold temperatures promote transcriptional shutdown of Arabidopsis FLOWERING LOCUS C (FLC), independently of factors conferring epigenetic memory. As winter continues, expression of VERNALIZATION INSENSITIVE3 (VIN3), a factor needed for epigenetic silencing, is upregulated by at least two independent thermosensory processes. One integrates long-term cold temperatures, while the other requires the absence of daily temperatures above 15 °C. The lack of spikes of high temperature, not just prolonged cold, is thus the major driver for vernalisation. Monitoring of peak daily temperature is an effective mechanism to judge seasonal progression, but is likely to have deleterious consequences for vernalisation as the climate becomes more variable.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/000C0625/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/000PR9773/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/000PR9788/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- ERC_/European Research Council/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

