Cotranslational protein assembly imposes evolutionary constraints on homomeric proteins
- PMID: 29434345
- PMCID: PMC5995306
- DOI: 10.1038/s41594-018-0029-5
Cotranslational protein assembly imposes evolutionary constraints on homomeric proteins
Abstract
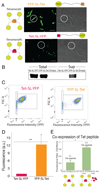
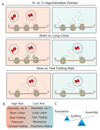
Cotranslational protein folding can facilitate rapid formation of functional structures. However, it can also cause premature assembly of protein complexes, if two interacting nascent chains are in close proximity. By analyzing known protein structures, we show that homomeric protein contacts are enriched toward the C termini of polypeptide chains across diverse proteomes. We hypothesize that this is the result of evolutionary constraints for folding to occur before assembly. Using high-throughput imaging of protein homomers in Escherichia coli and engineered protein constructs with N- and C-terminal oligomerization domains, we show that, indeed, proteins with C-terminal homomeric interface residues consistently assemble more efficiently than those with N-terminal interface residues. Using in vivo, in vitro and in silico experiments, we identify features that govern successful assembly of homomers, which have implications for protein design and expression optimization.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

