ChemTS: an efficient python library for de novo molecular generation
- PMID: 29435094
- PMCID: PMC5801530
- DOI: 10.1080/14686996.2017.1401424
ChemTS: an efficient python library for de novo molecular generation
Abstract
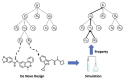
Automatic design of organic materials requires black-box optimization in a vast chemical space. In conventional molecular design algorithms, a molecule is built as a combination of predetermined fragments. Recently, deep neural network models such as variational autoencoders and recurrent neural networks (RNNs) are shown to be effective in de novo design of molecules without any predetermined fragments. This paper presents a novel Python library ChemTS that explores the chemical space by combining Monte Carlo tree search and an RNN. In a benchmarking problem of optimizing the octanol-water partition coefficient and synthesizability, our algorithm showed superior efficiency in finding high-scoring molecules. ChemTS is available at https://github.com/tsudalab/ChemTS.
Keywords: 404 Materials informatics / Genomics; 60 New topics/Others; Molecular design; Monte Carlo tree search; python library; recurrent neural network.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures


References
-
- Niu G, Guo X, Wang L. Review of recent progress in chemical stability of perovskite solar cells. J Mater Chem A. 2015;3(17):8970–8980.
-
- Ueda A, Yamada S, Isono T, et al. . Hydrogen-bond-dynamics-based switching of conductivity and magnetism: a phase transition caused by deuterium and electron transfer in a hydrogen-bonded purely organic conductor crystal. J Am Chem Soc. 2014;136(34):12184–12192. - PubMed
-
- Yeung MCL, Yam VWW. Luminescent cation sensors: from host-guest chemistry, supramolecular chemistry to reaction-based mechanisms. Chem Soc Rev. 2015;44(13):4192–4202. - PubMed
-
- Horiuchi S, Tokura Y. Organic ferroelectrics. Nat Mater. 2008;7(5):357–366. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
