The unique fold and lability of the [2Fe-2S] clusters of NEET proteins mediate their key functions in health and disease
- PMID: 29435647
- PMCID: PMC6006223
- DOI: 10.1007/s00775-018-1538-8
The unique fold and lability of the [2Fe-2S] clusters of NEET proteins mediate their key functions in health and disease
Abstract
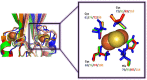
NEET proteins comprise a new class of [2Fe-2S] cluster proteins. In human, three genes encode for NEET proteins: cisd1 encodes mitoNEET (mNT), cisd2 encodes the Nutrient-deprivation autophagy factor-1 (NAF-1) and cisd3 encodes MiNT (Miner2). These recently discovered proteins play key roles in many processes related to normal metabolism and disease. Indeed, NEET proteins are involved in iron, Fe-S, and reactive oxygen homeostasis in cells and play an important role in regulating apoptosis and autophagy. mNT and NAF-1 are homodimeric and reside on the outer mitochondrial membrane. NAF-1 also resides in the membranes of the ER associated mitochondrial membranes (MAM) and the ER. MiNT is a monomer with distinct asymmetry in the molecular surfaces surrounding the clusters. Unlike its paralogs mNT and NAF-1, it resides within the mitochondria. NAF-1 and mNT share similar backbone folds to the plant homodimeric NEET protein (At-NEET), while MiNT's backbone fold resembles a bacterial MiNT protein. Despite the variation of amino acid composition among these proteins, all NEET proteins retained their unique CDGSH domain harboring their unique 3Cys:1His [2Fe-2S] cluster coordination through evolution. The coordinating exposed His was shown to convey the lability to the NEET proteins' [2Fe-2S] clusters. In this minireview, we discuss the NEET fold and its structural elements. Special attention is given to the unique lability of the NEETs' [2Fe-2S] cluster and the implication of the latter to the NEET proteins' cellular and systemic function in health and disease.
Keywords: Cisd(1–3) encoded NEET proteins; Iron-sulfur clusters; NEET-cluster lability; NEET-fold; [2Fe-2S].
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

