Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells
- PMID: 29436394
- PMCID: PMC5839763
- DOI: 10.1084/jem.20171626
Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells
Abstract
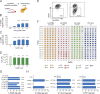
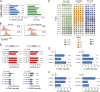
CRISPR (clustered, regularly interspaced, short palindromic repeats)/Cas9 (CRISPR-associated protein 9) has become the tool of choice for generating gene knockouts across a variety of species. The ability for efficient gene editing in primary T cells not only represents a valuable research tool to study gene function but also holds great promise for T cell-based immunotherapies, such as next-generation chimeric antigen receptor (CAR) T cells. Previous attempts to apply CRIPSR/Cas9 for gene editing in primary T cells have resulted in highly variable knockout efficiency and required T cell receptor (TCR) stimulation, thus largely precluding the study of genes involved in T cell activation or differentiation. Here, we describe an optimized approach for Cas9/RNP transfection of primary mouse and human T cells without TCR stimulation that results in near complete loss of target gene expression at the population level, mitigating the need for selection. We believe that this method will greatly extend the feasibly of target gene discovery and validation in primary T cells and simplify the gene editing process for next-generation immunotherapies.
© 2018 Genentech.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

