Differential expression profiles of long noncoding RNA and mRNA in colorectal cancer tissues from patients with lung metastasis
- PMID: 29436635
- PMCID: PMC5866008
- DOI: 10.3892/mmr.2018.8576
Differential expression profiles of long noncoding RNA and mRNA in colorectal cancer tissues from patients with lung metastasis
Abstract
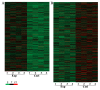
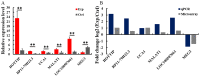
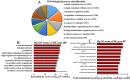
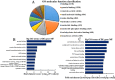
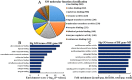
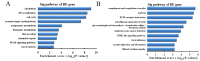
Lungs are the most common extra‑abdominal site of metastasis of colorectal cancer (CRC), in which long noncoding RNA (lncRNA) may serve a role. In the present study, a high‑throughput microarray assay was performed to detect lncRNA expression and identify novel targets for further study of lung metastasis in CRC. In the CRC tissues from patients with lung metastasis, 7,632 lncRNA (3,574 upregulated and 4,058 downregulated) and 6,185 mRNA (3,394 upregulated and 2,791 downregulated) were detected to be differentially expressed with a fold change ≥2 and P<0.05 compared with the CRC tissues without metastasis. A total of six differentially regulated lncRNA were confirmed by reverse transcription‑quantitative polymerase chain reaction in 20 pairs of CRC samples. Furthermore, gene ontology and pathway analysis were conducted to predict the possible roles of the identified mRNA. The upregulated mRNA were associated with cell division (biological processes), protein kinase B binding (molecular functions) and cellular components. The downregulated mRNA were associated with cell adhesion, platelet‑derived growth factor binding and membrane components. Pathway analysis determined that the upregulated mRNA were associated with the Wnt signaling pathway in the CRC tissues from patients with lung metastasis, while the downregulated mRNA were associated with the phosphoinositide 3‑kinase/Akt signaling pathway. The results of the present study suggested that differentially expressed lncRNA may be associated with lung metastasis and may provide insights into the biology and prevention of lung metastasis.
Keywords: colorectal cancer; lung metastasis; long noncoding RNA; biological process; molecular function; cellular component.
Figures




References
-
- Lin PC, Lin JK, Lin CC, Wang HS, Yang SH, Jiang JK, Lan YT, Lin TC, Li AF, Chen WS, Chang SC. Carbohydrate antigen 19-9 is a valuable prognostic factor in colorectal cancer patients with normal levels of carcinoembryonic antigen and may help predict lung metastasis. Int J Colorectal Dis. 2012;27:1333–1338. doi: 10.1007/s00384-012-1447-1. - DOI - PubMed
-
- Cejas P, López-Gómez M, Aguayo C, Madero R, de Castro Carpeño J, Belda-Iniesta C, Barriuso J, Moreno Garcia V, Larrauri J, López R, et al. KRAS mutations in primary colorectal cancer tumors and related metastases: A potential role in prediction of lung metastasis. PLoS One. 2009;4:e8199. doi: 10.1371/journal.pone.0008199. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

