DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus
- PMID: 29437559
- PMCID: PMC5909746
- DOI: 10.1136/annrheumdis-2017-212379
DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus
Abstract
Objectives: Systemic lupus erythematosus (SLE) is a chronic autoimmune condition with heterogeneous presentation and complex aetiology where DNA methylation changes are emerging as a contributing factor. In order to discover novel epigenetic associations and investigate their relationship to genetic risk for SLE, we analysed DNA methylation profiles in a large collection of patients with SLE and healthy individuals.
Methods: DNA extracted from blood from 548 patients with SLE and 587 healthy controls were analysed on the Illumina HumanMethylation 450 k BeadChip, which targets 485 000 CpG sites across the genome. Single nucleotide polymorphism (SNP) genotype data for 196 524 SNPs on the Illumina ImmunoChip from the same individuals were utilised for methylation quantitative trait loci (cis-meQTLs) analyses.
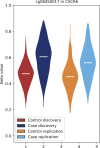
Results: We identified and replicated differentially methylated CpGs (DMCs) in SLE at 7245 CpG sites in the genome. The largest methylation differences were observed at type I interferon-regulated genes which exhibited decreased methylation in SLE. We mapped cis-meQTLs and identified genetic regulation of methylation levels at 466 of the DMCs in SLE. The meQTLs for DMCs in SLE were enriched for genetic association to SLE, and included seven SLE genome-wide association study (GWAS) loci: PTPRC (CD45), MHC-class III, UHRF1BP1, IRF5, IRF7, IKZF3 and UBE2L3. In addition, we observed association between genotype and variance of methylation at 20 DMCs in SLE, including at the HLA-DQB2 locus.
Conclusions: Our results suggest that several of the genetic risk variants for SLE may exert their influence on the phenotype through alteration of DNA methylation levels at regulatory regions of target genes.
Keywords: gene polymorphism; systemic lupus erythematosus.
© Article author(s) (or their employer(s) unless otherwise stated in the text of the article) 2018. All rights reserved. No commercial use is permitted unless otherwise expressly granted.
Conflict of interest statement
Competing interests: None declared.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

