Staphylococcus aureus HemX Modulates Glutamyl-tRNA Reductase Abundance To Regulate Heme Biosynthesis
- PMID: 29437922
- PMCID: PMC5801465
- DOI: 10.1128/mBio.02287-17
Staphylococcus aureus HemX Modulates Glutamyl-tRNA Reductase Abundance To Regulate Heme Biosynthesis
Abstract
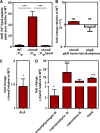
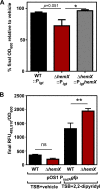
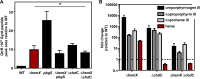
Staphylococcus aureus is responsible for a significant amount of devastating disease. Its ability to colonize the host and cause infection is supported by a variety of proteins that are dependent on the cofactor heme. Heme is a porphyrin used broadly across kingdoms and is synthesized de novo from common cellular precursors and iron. While heme is critical to bacterial physiology, it is also toxic in high concentrations, requiring that organisms encode regulatory processes to control heme homeostasis. In this work, we describe a posttranscriptional regulatory strategy in S. aureus heme biosynthesis. The first committed enzyme in the S. aureus heme biosynthetic pathway, glutamyl-tRNA reductase (GtrR), is regulated by heme abundance and the integral membrane protein HemX. GtrR abundance increases dramatically in response to heme deficiency, suggesting a mechanism by which S. aureus responds to the need to increase heme synthesis. Additionally, HemX is required to maintain low levels of GtrR in heme-proficient cells, and inactivation of hemX leads to increased heme synthesis. Excess heme synthesis in a ΔhemX mutant activates the staphylococcal heme stress response, suggesting that regulation of heme synthesis is critical to reduce self-imposed heme toxicity. Analysis of diverse organisms indicates that HemX is widely conserved among heme-synthesizing bacteria, suggesting that HemX is a common factor involved in the regulation of GtrR abundance. Together, this work demonstrates that S. aureus regulates heme synthesis by modulating GtrR abundance in response to heme deficiency and through the activity of the broadly conserved HemX.IMPORTANCEStaphylococcus aureus is a leading cause of skin and soft tissue infections, endocarditis, bacteremia, and osteomyelitis, making it a critical health care concern. Development of new antimicrobials against S. aureus requires knowledge of the physiology that supports this organism's pathogenesis. One component of staphylococcal physiology that contributes to growth and virulence is heme. Heme is a widely utilized cofactor that enables diverse chemical reactions across many enzyme families. S. aureus relies on many critical heme-dependent proteins and is sensitive to excess heme toxicity, suggesting S. aureus must maintain proper intracellular heme homeostasis. Because S. aureus provides heme for heme-dependent enzymes via synthesis from common precursors, we hypothesized that regulation of heme synthesis is one mechanism to maintain heme homeostasis. In this study, we identify that S. aureus posttranscriptionally regulates heme synthesis by restraining abundance of the first heme biosynthetic enzyme, GtrR, via heme and the broadly conserved membrane protein HemX.
Keywords: Staphylococcus aureus; heme; tetrapyrroles.
Copyright © 2018 Choby et al.
Figures




References
-
- Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core surveillance (ABCs) MRSA Investigators . 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

