Induction of Genotype Cross-Reactive, Hepatitis C Virus-Specific, Cell-Mediated Immunity in DNA-Vaccinated Mice
- PMID: 29437963
- PMCID: PMC5874410
- DOI: 10.1128/JVI.02133-17
Induction of Genotype Cross-Reactive, Hepatitis C Virus-Specific, Cell-Mediated Immunity in DNA-Vaccinated Mice
Abstract
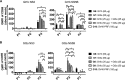
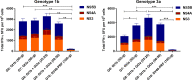
A universal hepatitis C virus (HCV) vaccine should elicit multiantigenic, multigenotypic responses, which are more likely to protect against challenge with the range of genotypes and subtypes circulating in the community. A vaccine cocktail and vaccines encoding consensus HCV sequences are attractive approaches to achieve this goal. Consequently, in a series of mouse vaccination studies, we compared the immunogenicity of a DNA vaccine encoding a consensus HCV nonstructural 5B (NS5B) protein to that of a cocktail of DNA plasmids encoding the genotype 1b (Gt1b) and Gt3a NS5B proteins. To complement this study, we assessed responses to a multiantigenic cocktail regimen by comparing a DNA vaccine cocktail encoding Gt1b and Gt3a NS3, NS4, and NS5B proteins to a single-genotype NS3/4/5B DNA vaccine. To thoroughly evaluate in vivo cytotoxic T lymphocyte (CTL) and T helper (Th) cell responses against Gt1b and Gt3a HCV peptide-pulsed target cells, we exploited a novel fluorescent-target array (FTA). FTA and enzyme-linked immunosorbent spot (ELISpot) analyses collectively indicated that the cocktail regimens elicited higher responses to Gt1b and Gt3a NS5B proteins than those with the consensus vaccine, while the multiantigenic DNA cocktail significantly increased the responses to NS3 and NS5B compared to those elicited by the single-genotype vaccines. Thus, a DNA cocktail vaccination regimen is more effective than a consensus vaccine or a monovalent vaccine at increasing the breadth of multigenotypic T cell responses, which has implications for the development of vaccines for communities where multiple HCV genotypes circulate.IMPORTANCE Despite the development of highly effective direct-acting antivirals (DAA), infections with hepatitis C virus (HCV) continue, particularly in countries where the supply of DAA is limited. Furthermore, patients who eliminate the virus as a result of DAA therapy can still be reinfected. Thus, a vaccine for HCV is urgently required, but the heterogeneity of HCV strains makes the development of a universal vaccine difficult. To address this, we developed a novel cytolytic DNA vaccine which elicits robust cell-mediated immunity (CMI) to the nonstructural (NS) proteins in vaccinated animals. We compared the immune responses against genotypes 1 and 3 that were elicited by a consensus DNA vaccine or a DNA vaccine cocktail and showed that the cocktail induced higher levels of CMI to the NS proteins of both genotypes. This study suggests that a universal HCV vaccine can most readily be achieved by use of a DNA vaccine cocktail.
Keywords: DNA vaccines; ELISpot; HCV; NS5; consensus; hepatitis; multiantigenic; multigenotypic; vaccine.
Copyright © 2018 American Society for Microbiology.
Figures




References
-
- WHO. 2017. Global hepatitis report. WHO, Geneva, Switzerland.
-
- Yu MY, Bartosch B, Zhang P, Guo ZP, Renzi PM, Shen LM, Granier C, Feinstone SM, Cosset FL, Purcell RH. 2004. Neutralizing antibodies to hepatitis C virus (HCV) in immune globulins derived from anti-HCV-positive plasma. Proc Natl Acad Sci U S A 101:7705–7710. doi: 10.1073/pnas.0402458101. - DOI - PMC - PubMed
-
- Broering TJ, Garrity KA, Boatright NK, Sloan SE, Sandor F, Thomas WD Jr, Szabo G, Finberg RW, Ambrosino DM, Babcock GJ. 2009. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J Virol 83:12473–12482. doi: 10.1128/JVI.01138-09. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

