Herpes Simplex Virus 1 Dramatically Alters Loading and Positioning of RNA Polymerase II on Host Genes Early in Infection
- PMID: 29437966
- PMCID: PMC5874419
- DOI: 10.1128/JVI.02184-17
Herpes Simplex Virus 1 Dramatically Alters Loading and Positioning of RNA Polymerase II on Host Genes Early in Infection
Abstract
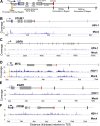
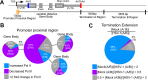
Herpes simplex virus 1 (HSV-1) transcription is mediated by cellular RNA polymerase II (Pol II). Recent studies investigating how Pol II transcription of host genes is altered after HSV-1 are conflicting. Chromatin immunoprecipitation sequencing (ChIP-seq) studies suggest that Pol II is almost completely removed from host genes at 4 h postinfection (hpi), while 4-thiouridine (4SU) labeling experiments show that host transcription termination is extended at 7 hpi, implying that a significant amount of Pol II remains associated with host genes in infected cells. To address this discrepancy, we used precision nuclear run-on analysis (PRO-seq) to determine the location of Pol II to single-base-pair resolution in combination with quantitative reverse transcription-PCR (qRT-PCR) analysis at 3 hpi. HSV-1 decreased Pol II on approximately two-thirds of cellular genes but increased Pol II on others. For more than 85% of genes for which transcriptional termination could be statistically assessed, Pol II was displaced to positions downstream of the normal termination zone, suggesting extensive termination defects. Pol II amounts at the promoter, promoter-proximal pause site, and gene body were also modulated in a gene-specific manner. qRT-PCR of selected RNAs showed that HSV-1-induced extension of the termination zone strongly correlated with decreased RNA and mRNA accumulation. However, HSV-1-induced increases of Pol II occupancy on genes without termination zone extension correlated with increased cytoplasmic mRNA. Functional grouping of genes with increased Pol II occupancy suggested an upregulation of exosome secretion and downregulation of apoptosis, both of which are potentially beneficial to virus production.IMPORTANCE This study provides a map of RNA polymerase II location on host genes after infection with HSV-1 with greater detail than previous ChIP-seq studies and rectifies discrepancies between ChIP-seq data and 4SU labeling experiments with HSV-1. The data show the effects that a given change in RNA Pol II location on host genes has on the abundance of different RNA types, including nuclear, polyadenylated mRNA and cytoplasmic, polyadenylated mRNA. It gives a clearer understanding of how HSV-1 augments host transcription of some genes to provide an environment favorable to HSV-1 replication.
Keywords: RNA polymerases; herpes simplex virus; human herpesviruses; transcriptional regulation; transcriptional repression.
Copyright © 2018 American Society for Microbiology.
Figures



References
-
- Whitley R, Kimberlin DW, Prober CG. 2007. HSV-1 and 2: pathogenesis and disease, p 589–601. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. - PubMed
-
- Carey M, Peterson CL, Smale ST. 2009. Transcriptional regulation in eukaryotes: concepts, strategies, and techniques, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

