N-Terminomics TAILS Identifies Host Cell Substrates of Poliovirus and Coxsackievirus B3 3C Proteinases That Modulate Virus Infection
- PMID: 29437971
- PMCID: PMC5874412
- DOI: 10.1128/JVI.02211-17
N-Terminomics TAILS Identifies Host Cell Substrates of Poliovirus and Coxsackievirus B3 3C Proteinases That Modulate Virus Infection
Abstract
Enteroviruses encode proteinases that are essential for processing of the translated viral polyprotein. In addition, viral proteinases also target host proteins to manipulate cellular processes and evade innate antiviral responses to promote replication and infection. Although some host protein substrates of enterovirus proteinases have been identified, the full repertoire of targets remains unknown. We used a novel quantitative in vitro proteomics-based approach, termed
Keywords: RNA replication; coxsackievirus; enterovirus; plus-strand RNA virus; poliovirus; proteases; proteinase; proteomics.
Copyright © 2018 Jagdeo et al.
Figures

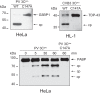
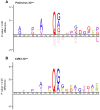


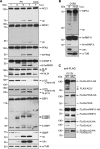


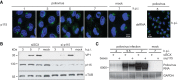


References
-
- Marchant D, Si X, Luo H, McManus B, Yang D. 2008. The impact of CVB3 infection on host cell biology. Curr Top Microbiol Immunol 323:177–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

