Reduction of Hexavalent Chromium and Detection of Chromate Reductase (ChrR) in Stenotrophomonas maltophilia
- PMID: 29438314
- PMCID: PMC6017488
- DOI: 10.3390/molecules23020406
Reduction of Hexavalent Chromium and Detection of Chromate Reductase (ChrR) in Stenotrophomonas maltophilia
Abstract
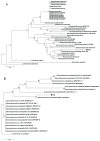
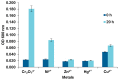
An Gram negative strain of S. maltophilia, indigenous to environments contaminated by Cr(VI) and identified by biochemical methods and 16S rRNA gene analysis, reduced chromate by 100%, 98-99% and 92% at concentrations in the 10-70, 80-300, and 500 mg/L range, respectively at pH 7 and temperature 37 °C. Increasing concentrations of Cr(VI) in the medium lowered the growth rate but could not be directly correlated with the amount of Cr(VI) reduced. The strain also exhibited multiple resistance to antibiotics and tolerance and resistance to various heavy metals (Ni, Zn and Cu), with the exception of Hg. Hexavalent chromium reduction was mainly associated with the soluble fraction of the cell evaluated with crude cell-free extracts. A protein of molecular weight around 25 kDa was detected on SDS-PAGE gel depending on the concentration of hexavalent chromium in the medium (0, 100 and 500 mg/L). In silico analysis in this contribution, revealed the presence of the chromate reductase gene ChrR in S. maltophilia, evidenced through a fragment of around 468 bp obtained experimentally. High Cr(VI) concentration resistance and high Cr(VI) reducing ability of the strain make it a suitable candidate for bioremediation.
Keywords: ChrR; Cr(VI); Stenotrophomonas maltophilia; chromate reductase; heavy metal.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Reduction of chromium-VI by chromium-resistant Escherichia coli FACU: a prospective bacterium for bioremediation.Folia Microbiol (Praha). 2020 Aug;65(4):687-696. doi: 10.1007/s12223-020-00771-y. Epub 2020 Jan 27. Folia Microbiol (Praha). 2020. PMID: 31989423
-
Reduction of chromate by cell-free extract of Brucella sp. isolated from Cr(VI) contaminated sites.Bioresour Technol. 2007 May;98(8):1541-7. doi: 10.1016/j.biortech.2006.06.011. Epub 2006 Aug 22. Bioresour Technol. 2007. PMID: 16931000
-
Hexavalent Chromium Reduction by Microbacterium oleivorans A1: A Possible Mechanism of Chromate -Detoxification and -Bioremediation.Recent Pat Biotechnol. 2016;9(2):116-29. doi: 10.2174/187220830902160308192126. Recent Pat Biotechnol. 2016. PMID: 26961669
-
Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review.J Environ Manage. 2014 Dec 15;146:383-399. doi: 10.1016/j.jenvman.2014.07.014. Epub 2014 Sep 8. J Environ Manage. 2014. PMID: 25199606 Review.
-
Mechanisms of hexavalent chromium resistance and removal by microorganisms.Rev Environ Contam Toxicol. 2015;233:45-69. doi: 10.1007/978-3-319-10479-9_2. Rev Environ Contam Toxicol. 2015. PMID: 25367133 Review.
Cited by
-
Characterization and Molecular Insights of a Chromium-Reducing Bacterium Bacillus tropicus.Microorganisms. 2024 Dec 19;12(12):2633. doi: 10.3390/microorganisms12122633. Microorganisms. 2024. PMID: 39770835 Free PMC article.
-
Biodegradation of Polypropylene in Presence of Chromium Mediated by Stenotrophomonas sp. and Lysinibacillus sp. Isolated from Wetland Sediments.Curr Microbiol. 2025 Jul 7;82(8):369. doi: 10.1007/s00284-025-04353-4. Curr Microbiol. 2025. PMID: 40624338
-
Isolation of Aerobic Heterotrophic Bacteria from a Microbial Mat with the Ability to Grow on and Remove Hexavalent Chromium through Biosorption and Bioreduction.Appl Biochem Biotechnol. 2025 Jan;197(1):94-112. doi: 10.1007/s12010-024-05023-0. Epub 2024 Aug 5. Appl Biochem Biotechnol. 2025. PMID: 39102082
-
Tolerance and reduction of chromium by bacterial strains.Arch Microbiol. 2022 Nov 25;204(12):730. doi: 10.1007/s00203-022-03329-3. Arch Microbiol. 2022. PMID: 36434407
-
Efficacy of Lacticaseibacillus rhamnosus probiotic strains in treating chromate induced dermatitis.Sci Rep. 2025 Mar 14;15(1):8796. doi: 10.1038/s41598-025-93732-9. Sci Rep. 2025. PMID: 40087372 Free PMC article.
References
-
- Chattopadhyay B., Utpal S.R., Mukhopadhyay S. Mobility and bioavailability of chromium in the environment: Physico-chemical and microbial oxidation of Cr (III) to Cr (VI) J. Appl. Sci. Environ. Manag. 2010;14:97–101. doi: 10.4314/jasem.v14i2.57873. - DOI
-
- Cheung K.H., Ji-Dong G.U. Mechanism of hexavalent chromium detoxification by microorganisms and bioremediation application potential: A review. Int. Biodeterior. Biodegrad. 2007;59:8–15. doi: 10.1016/j.ibiod.2006.05.002. - DOI
-
- Maqbool Z., Asghar H.N., Shahzad T., Hussain S., Riaz M., Ali S., Maqsood M. Isolating, screening and applying chromium reducing bacteria to promote growth and yield of okra (Hibiscus esculentus L.) in chromium contaminated soils. Ecotoxicol. Environ. Saf. 2015;114:343–349. doi: 10.1016/j.ecoenv.2014.07.007. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

