Diurnal transcriptome atlas of a primate across major neural and peripheral tissues
- PMID: 29439024
- PMCID: PMC5924732
- DOI: 10.1126/science.aao0318
Diurnal transcriptome atlas of a primate across major neural and peripheral tissues
Abstract
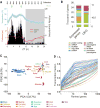
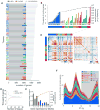
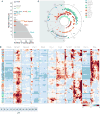
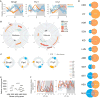
Diurnal gene expression patterns underlie time-of-the-day-specific functional specialization of tissues. However, available circadian gene expression atlases of a few organs are largely from nocturnal vertebrates. We report the diurnal transcriptome of 64 tissues, including 22 brain regions, sampled every 2 hours over 24 hours, from the primate Papio anubis (baboon). Genomic transcription was highly rhythmic, with up to 81.7% of protein-coding genes showing daily rhythms in expression. In addition to tissue-specific gene expression, the rhythmic transcriptome imparts another layer of functional specialization. Most ubiquitously expressed genes that participate in essential cellular functions exhibit rhythmic expression in a tissue-specific manner. The peak phases of rhythmic gene expression clustered around dawn and dusk, with a "quiescent period" during early night. Our findings also unveil a different temporal organization of central and peripheral tissues between diurnal and nocturnal animals.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures



Comment in
-
Rhythms: The dark side meets the light.Science. 2018 Mar 16;359(6381):1210-1211. doi: 10.1126/science.aat3211. Science. 2018. PMID: 29590060 No abstract available.
-
Circadian clock: each tissue has its own rhythm.Cardiovasc Res. 2018 Jul 1;114(8):e57-e59. doi: 10.1093/cvr/cvy098. Cardiovasc Res. 2018. PMID: 30052895 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

