Programming gene and engineered-cell therapies with synthetic biology
- PMID: 29439214
- PMCID: PMC7643872
- DOI: 10.1126/science.aad1067
Programming gene and engineered-cell therapies with synthetic biology
Abstract
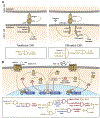
Gene and engineered-cell therapies promise to treat diseases by genetically modifying cells to carry out therapeutic tasks. Although the field has had some success in treating monogenic disorders and hematological malignancies, current approaches are limited to overexpression of one or a few transgenes, constraining the diseases that can be treated with this approach and leading to potential concerns over safety and efficacy. Synthetic gene networks can regulate the dosage, timing, and localization of gene expression and therapeutic activity in response to small molecules and disease biomarkers. Such "programmable" gene and engineered-cell therapies will provide new interventions for incurable or difficult-to-treat diseases.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

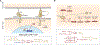
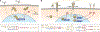
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

